Back to Journals » Journal of Asthma and Allergy » Volume 14
LncRNA H19 Inhibits Proliferation and Migration of Airway Smooth Muscle Cells Induced by PDGF-BB Through miR-21/PTEN/Akt Axis
Received 13 November 2020
Accepted for publication 18 January 2021
Published 28 January 2021 Volume 2021:14 Pages 71—80
DOI https://doi.org/10.2147/JAA.S291333
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Amrita Dosanjh
Haiying Yu, Ningning Qi, Qingxia Zhou
Department of Pediatrics, Weifang People’s Hospital, Weifang, Shandong 261041, People’s Republic of China
Correspondence: Qingxia Zhou
Department of Pediatrics, Weifang People’s Hospital, Guangwen Street, Kuiwen District, Wenfang, Shandong 261041, People’s Republic of China
Tel +86-05368248176
Email [email protected]
Background: LncRNA H19 expression is down-regulated in patients with asthma. The hyperplasia of airway smooth muscle cells (ASMCs) promotes the development of airway remodeling in asthma. Therefore, we attempted to evaluate the regulatory function of H19 in the proliferation and migration of ASMCs.
Methods: The expressions of H19 and miR-21 were detected using qRT-PCR. PDGF-BB-induced abnormal proliferation and migration of ASMCs was used as the airway remodeling model in vitro. The expressions of H19 and miR-21 were modified by transfection with pcDNA3.1-H19 and miR-21 mimic, respectively. CCK-8 assay, flow cytometry-based cell cycle analysis was conducted to examine the proliferation ability of ASMCs. The migration ability was measured by transwell assay. Dual-luciferase reporter system was carried out to find the potential relationship between miR-21 and H19 or PTEN. Western blot was conducted to detect the expressions of PCNA, MMP-9, α-SMA, PTEN, and the phosphorylation level of Akt.
Results: LncRNA-H19 expression was decreased and microRNA-21 expression was increased in serum samples of children with asthma and PDGF-BB-stimulated ASMCs. Overexpression of H19 reduced the proliferation and migration ability of ASMCs with PDGF-BB treatment and these changes were reversed by miR-21 mimic. H19 promoted the protein level of PTEN via sponging miR-21. Overexpression of H19 suppressed miR-21-induced phosphorylation of Akt, and the suppression effect of H19 on phosphorylation of Akt was significantly reduced after transfecting shPTEN in ASMCs.
Conclusion: In this study, overexpression of H19 suppressed the proliferation and migration of ASMCs induced by PDGF-BB via miR-21/PTEN/Akt axis, which could be a potential biomarker and target for treating hyperplasia of airway smooth muscle cells.
Keywords: lncRNA H19, asthmatic children, airway smooth muscle cell, miR-21, airway remodeling
Introduction
Asthma in children is the most common chronic respiratory disease worldwide,1 which significantly affects children’s life quality and causes increasing hospitalization and economic burden.2,3 Airway remodeling is a pathological character of asthma.4 Current therapeutics, such as anti-inflammation agents, can ameliorate airway inflammation. However, there is no available method to reverse airway remodeling. Recently increasing researches revealed that airway smooth muscle cells (ASMCs) proliferated and migrated abnormally to promote the development of airway remodeling.5,6 The phenotypic transformation of ASMCs acts as a key mediator in cell proliferation of airway remodeling.7 When stimulated by platelet-derived growth factor-BB (PDGF-BB), the contraction phenotype of ASMCs could transform to synthesis phenotype.8 Synthetic ASMCs can secrete cytokines, growth factors, matrix protein or cellular adhesion molecules, such as IL-1, IL-8, TGF-β1, collagen Ι and fibronectin, and abnormally proliferate to promote the hyperplasia and thickness of the airway smooth muscle layer.9,10 Taken together, the study to investigate related genes in ASMCs proliferation and migration was of great significance for finding potential biomarkers for early prevention and intervention of asthma in children.
Long non-coding RNA (lncRNA) composes of more than 200 nucleotides, regulates gene expression and thus takes effect in the initiation of the pathological process in many diseases.11 Previous researches indicated that lncRNA could regulate the proliferation and migration ability of ASMCs in asthma.12,13 Recent studies found that lncRNA-H19 expression was remarkedly reduced in patients with asthma.12,14 However, the influence of H19 on asthma remains unclear.
LncRNA could directly sponge specific microRNA to regulate microRNA target gene expression and affect biological conditions, calling a competitive endogenous RNA network (ceRNET). The expressions of several microRNAs were dysregulated in patients with asthma, such as miR-146a, miR-21, and miR-485-3p.15,16 Of these, miR-21 expression was up-regulated and it might serve as a better biomarker in asthmatic children.15 And miR-21 could promote the proliferation and migration of ASMCs.17 A recent report revealed that H19 regulated inflammation and nerve injury via directly sponging miR-21 to promote the expression of PDCA4 in retinal ischemia/reperfusion injury.18 However, whether miR-21 could be regulated by H19 in children with asthma remains to be investigated.
The PI3K/Akt signaling pathway is necessary for airway remodeling of asthma.19 Noteworthy, miR-21 could decrease the expression of PTEN, the down-regulator of Akt, and then promoted the pathological change of ASMCs.20,21 In this study, we aimed to find the interaction between lncRNA-H19 and miR-21 in children with asthma. Furthermore, we investigated whether H19 played a role in ASMCs stimulated by PDGF-BB through PI3K/Akt pathway.
Materials and Methods
Serum Samples Collection
A total of 60 children diagnosed with asthma and 28 healthy volunteers in Weifang People’s Hospital from May 2016 to June 2018 were included in this study. Approval was obtained from the Ethics Committee of Weifang People’s Hospital. Informed consent of all children’s parents was attained. We confirmed this study was conducted in accordance with the Declaration of Helsinki. Children with asthma were diagnosed according to the revised version of the Chinese guideline for the prevention and management of bronchial asthma. The clinical information of all subjects is shown in Table 1.
 |
Table 1 Clinical Information of Subjects |
Cell Culture
The human airway smooth muscle cell (hASMC) was obtained from Bnbio (Beijing, China). HASMCs were cultured as described with the protocol provided. For the proliferation model in vitro, PDGF-BB (10 ng/mL) was used for the treatment of ASMCs for 24 h continuously.
Cell Transfection
Full-length H19 cDNA was obtained by PCR. And the sequence was sub-cloned into pcDNA-3.1 vectors (Sangon Biotech, China), and the sequence of the recombinant vector was verified. The miR-21 mimic and corresponding control mimic (mimic NC) was synthesized by Shanghai GenePharm (Shanghai, China). Cells were transfected or co-transfected with Lipofectamine 3000 system (Invitrogen, USA). After transfection for 48 h, cells were collected for the following assays.
Quantitative Real-Time PCR Analysis
Total RNA was extracted from the clinical serum samples and ASMCs using TRIzol solution (Invitrogen, USA), and reverse-transcribed into cDNA. The expressions of the indicated genes were measured with Applied Biosystems with an SYBR mix. The sequences of specific primers used were as follows: H19 (F: 5ʹ-GCAGGTAGAGCGAGTAGCTG-3ʹ, and R: 5ʹ-TAGAGGCT TGGCTCCAGGAT-3ʹ); miR-21 (F: 5ʹ-GTCGTATCCAGTGCAGGGTCCGAGG TA-3ʹ, and R 5ʹ-CGCGCTAGCTTATCAGACTGA-3ʹ); PTEN (F: 5ʹ-TGGAT TCGACTTAGACTTGACCT-3ʹ, and R: 5ʹ-GGTGGGTTATGGTCTTCAAAAGG-3ʹ); U6 (F: 5ʹ- CTCGCTTCGGCAGCACA-3ʹ, and R: 5ʹ-AACGCTTCACGAATTTG CGT-3ʹ); GAPDH (F: 5ʹ-ACAACTTTGGTATCGTGGAAGG-3ʹ, and R: 5ʹ-GCCA TCACGCCACAGTTTC-3ʹ). U6 small noncoding RNA and GAPDH were applied as reference for miRNA and mRNA, respectively.
Western Blot Assay
HASMCs were subjected to extract protein with RIPA lysis buffer. BCA Protein Assay Kit (Thermo Scientific, USA) was applied for quantification of supernatant solution concentration. 20 ug proteins were separated using 10% SDS-PAGE gels electrophoresis, followed by PVDF membranes (Millipore, USA) transfer. After blocked with silk milk, the membranes were incubated with the following primary antibodies against PCNA (1:1500), MMP-9 (1:1100), α-SMA (1:1000), PTEN (1:900), Akt (1:1400), p-Akt (1:1200); GAPDH (1:4000) overnight at 4 °C. All of the primary antibodies were from Cell Signaling Technology (USA). Then, the membranes were incubated with 1:5000 diluted secondary antibodies. Finally, the expression levels of these proteins were visualized using ECL (Thermo Scientific, USA). The intensities of the bands were analyzed by ImageJ.
Cell Proliferation Analysis
The proliferation ability of ASMCs was measured with Cell Counting Kit (Beyotime, China). Three thousand ASMCs were added into each well of 48-well plates. Each group included three replicates. Then, 10 ul CCK-8 reagent was applied to incubate the cells for 12, 24, 48, 72 hours, respectively. After incubation for 3 hours at 37 °C, the cell viability was detected.
Cell Cycle Assay
70% ethanol was applied for the fixation of ASMCs. After washed twice using pre-cooled PBS, propidium iodide (PI, 50 ug/mL) and RNase Ι (250 ug/mL) were used to stain cells. The percentages of cell populations in different phases were analyzed using flow cytometry (Beckman Coulter, USA).
Transwell Assay
Transwell chamber with an 8-um pore size (Corning, USA) was applied to evaluate the migration ability of ASMCs. Cells were resuspended with serum-free DMEM and then seeded into the upper transwell chamber. The lower compartment was filled with DMEM containing 8% FBS with the stimulation of PDGF-BB or not. After 24 h incubation, non-migrated ASMCs that remained on the upper surface of the membrane were wiped off. Methanol was used for the fixation of cells on the lower surface. And then 0.1% crystal violet was used to stain the cells. Five random optical fields were photographed with a light microscope.
Dual-Luciferase Reporter Assay
To construct dual-luciferase reporter vectors, the predicted sequences of miR-21 binding sites in H19 (H19-WT) and PTEN (PTEN-WT) and their mutated binding sequences (H19-MUT and PTEN-MUT) were cloned into pGL-3 basic vector, respectively (Progma, USA). For luciferase assay, the recombinant vectors were co-transfected with miR-21 mimic or mimic-NC. Renilla reporter system was transfected into each group to normalize the transfection efficiency.
Statistical Analysis
Mean±standard deviation (SD) was analyzed by SPSS 20.0. The statistical analysis was executed by unpaired Student’s t-test to compare the two groups and one-way ANOVA with Dunnett’s post hoc test to compare three or more groups. When the p-value <0.05, the difference was considered significant. Each experiment was performed in three independent replicates.
Results
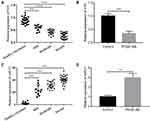
LncRNA H19 Expression Was Down-Regulated in Children with Asthma
To explore the effect of lncRNA H19 on children with asthma, we firstly detected the expression of H19 in serum samples from 60 asthmatic children and 28 healthy volunteers via qRT-PCR. We found that the expression level of H19 was decreased dramatically in the serum samples from asthmatic children compared with those from healthy volunteers and the expression level of H19 was decreased with increasing the severity of asthma (Figure 1A). Previous studies had shown that PDGF-BB induced the synthetic phenotype transformation of ASMCs, and synthetic phenotype ASMCs could proliferate and migrate abnormally.8 Thus, we also measured the expression level of H19 in ASMCs treated with PDGF-BB. Similarly, H19 expression was remarkably lower in cells with PDGF-BB treatment than those in cells without PDGF-BB treatment (Figure 1B). Then, the expression of miR-21 was evaluated. MiR-21 expression level was remarkably increased in asthmatic children and PDGF-BB stimulated ASMCs (Figure 1C and D). These results indicated that lncRNA H19 was associated with the progression of asthma.
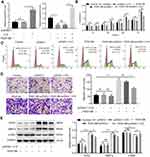
Overexpression of H19 Inhibited Proliferation and Migration Ability of ASMCs Induced by PDGF-BB
The hyperplasia of the airway smooth muscle layer is the key pathological character in asthma. The ASMCs with or without PDGF-BB treatment were transfected with pcDNA-3.1-H19 to measure the influence of H19 on ASMCs. The H19 expression was detected by qRT-PCR to ensure the transfection efficiency (Figure 2A). As shown in Figure 2B, the results of CCK-8 analysis showed that in PDGF-BB + pcDNA3.1-H19 group, H19 overexpression reduced the proliferation ability of PDGF-BB-stimulated ASMCs in contrast with the PDGF-BB + pcDNA3.1 group. Correspondingly, the results of flow cytometry showed that H19 overexpression arrested cell cycle in G0/G1 phase in PDGF-BB-stimulated ASMCs (Figure 2C). Meanwhile, as shown in Figure 2D, H19 suppressed the migration ability of ASMCs stimulated by PDGF-BB. We also assessed the expressions of proliferation marker PCNA, contractile phenotype biomarker α-SMA and MMP-9 via Western blot. The data revealed that H19 overexpression dramatically inhibited the expressions of PCNA and MMP-9, and promoted α-SMA expression in PDGF-BB-stimulated ASMCs (Figure 2E). And transfection with pcDNA3.1-H19 in ASMCs untreated with PDGF-BB arrested cell cycle in G0/G1 phase but made no significant change in proliferation and migration of ASMCs (Figure 2B–E). Taken all results above, H19 overexpression inhibited the proliferation and migration of ASMCs induced by PDGF-BB.
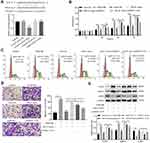
H19 Directly Targeted miR-21 and H19 Overexpression Markedly Reduced the Proliferation and Migration Ability of ASMCs Promoted by miR-21
To further elucidate the interaction between H19 and miR-21, we predicted the binding sites of miR-21 in H19 by bioinformatic analysis. As shown in Figure 3A, compared with co-transfection with mimic-NC and H19-WT, co-transfection with miR-21 mimic and wide type-H19 (H19-WT) significantly suppressed the luciferase activity of ASMCs, but luciferase activity was not changed in ASMCs transfected with mutated H19 reporter vector (H19-MUT), suggesting that miR-21 was a direct target of H19. Then, CCK-8 assay and cell cycle assay were carried out to measure whether H19 affected the proliferation ability of ASMCs with miR-21 overexpression. MiR-21 promoted the proliferation of ASMCs. But in comparison to miR-21 mimic + pcDNA3.1 group, the proliferation ability was decreased in miR-21 mimic + pcDNA3.1-H19 group (Figure 3B and C). Furthermore, the migration ability of ASMCs was evaluated with transwell assay. As shown in Figure 3D, the migrated cell number was remarkably decreased in miR-21 mimic + pcDNA3.1-H19 group in contrast with miR-21 mimic + pcDNA3.1 group. Similarly, H19 significantly abrogated the influence of miR-21 on the expression of PCNA, MMP-9 and α-SMA (Figure 3E). Collectively, these data demonstrated that H19 could suppress the proliferation and migration ability of ASMCs by directly sponging miR-21.
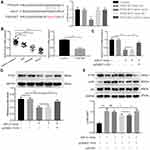
H19 Modulated PTEN Expression and Inhibited PI3K/Akt Signaling Pathway via Sponging miR-21
MiR-21 could decrease phosphatase and tensin homolog gene (PTEN) expression in asthma.17 Then, the dual-luciferase reporter system was applied to detect whether miR-21 could bind with PTEN 3ʹ-UTR. As shown in Figure 4A, transfection of miR-21 mimic decreased the luciferase activity of reporter system transfected with PTEN-WT compared with mimic-NC + PTEN-WT group. And miR-21 mimic had no influence on luciferase activity of cells with PTEN-MUT transfection. And we found that PTEN was downregulated in the serum of asthmatic children and PDGF-BB-stimulated ASMCs via qRT-PCR (Figure 4B). To further evaluate whether H19 regulates the expression of PTEN by sponging miR-21, we conducted qRT-PCR to detect the mRNA level and Western blot to evaluate the protein level. PTEN expression was declined in miR-21 mimic and miR-21 + pcDNA3.1 group, but remarkably increased in miR-21 mimic + pcDNA3.1-H19 group (Figure 4C and D). PTEN could down-regulate the phosphorylation level of Akt, which could subsequently suppress cell proliferation and migration. Therefore, we measured the phosphorylation level of Akt. We found that microRNA-21 mimic increased the phosphorylated form of Akt, but H19 distinctly abrogated the effect induced by miR-21 mimic (Figure 4E). And after knocking down PTEN in ASMCs co-transfected with pcDNA3.1-H19 and miR-21 mimic, the phosphorylation level of Akt was significantly elevated (Figure 4E). In conclusion, lncRNA H19 could inhibit the proliferation and migration of ASMC via miR-21/PTEN/Akt axis in PDGF-BB-stimulated ASMCs.
Discussion
Pathologic characters of asthma, including airway smooth muscle cells hyperplasia and subepithelial fibrosis, are associated with airway remodeling.22–24 And airway remodeling serves as an important influence on the development of asthma. When stimulated by PDGF-BB, the contractile phenotype of ASMCs can transform to the synthetic phenotype.8 The synthetic ASMCs could proliferate and migrate abnormally to promote the increasing thickness of the smooth muscle layer in airway remodeling. Therefore, in this study, ASMCs stimulated by PDGF-BB were used to be the proliferation model in vitro.
LncRNAs have been reported to emerge as key mediators in the pathological development of various respiratory diseases including asthma.25 For example, GAS5 promoted the proliferation of ASMCs via regulating miR-10a/BDNF signaling pathway.26 LncRNA PVT1 was associated with the aberrant proliferation of ASMCs.12 Recently, a study found that the expression of H19, a paternally imprinted and maternally expressed gene, was decreased in asthmatic patients compared with healthy people using microarray.12 Our results indicated that H19 expression was remarkably decreased in serum samples from children with asthma compared with samples of healthy volunteers and even in PDGF-BB stimulation ASMCs.
More researches focused on the regulatory role of H19 on tumorigenesis and inflammation so far. Su et al found that H19 could promote pulmonary artery smooth muscle cells to proliferate in PAH induced by MCT via sponging let-7b.27 In lung cancer, H19 promoted proliferation and metastasis of cancer cells via sponging miR-200a.28 These results demonstrated that H19 was closely associated with the enhancement of cell proliferation and migration. However, the regulatory influence of H19 on asthma is still indistinct.
In the present study, H19 suppressed cell proliferation and decreased the migration of ASMCs induced by PDGF-BB. And the expressions of PCNA and MMP-9 were significantly inhibited, and the contractile phenotype related protein α-SMA expression was increased after overexpression of H19 in ASMCs with PDGF-BB treatment. Therefore, we can conclude that H19 plays an inhibitory influence on the proliferation and migration ability of PDGF-BB-induced ASMCs.
A recent research elucidated that H19 initiated I/R-induced inflammation and neuronal lesion of retinal ischemia/reperfusion injury via directly sponging miR-21 in vivo.18 MiR-21 expression was prominently up-regulated in serum samples from asthmatic children.15 Moreover, miR-21 could enhance the cell number of proliferated or migrated ASMCs and contributed to airway remodeling of asthma.17 In our research, H19 could directly sponge miR-21 in ASMCs. Furthermore, H19 abrogated the enhanced influences of miR-21 on proliferation and the migration of ASMCs, indicating that H19 could inhibit the abnormal proliferation and migration of ASMCs via sponging miR-21.
Increasing evidences demonstrated that PI3K/Akt signaling pathway was associated with ASMCs proliferation and migration in asthma. Shao et al have shown that miR-133a regulated the expression of α-SMA via the PI3K/Akt pathway, which could promote airway remodeling of asthma.19 Galectin-1, a glycan-binding protein, exerted a suppression influence on proliferation, migration and phenotype switching of ASMCs stimulated by PDGF-BB through suppressing the activation of PI3K/Akt signaling pathway.29 Importantly, miR-21 could directly decrease the expression of PTEN in ASMCs.17
Our present study showed that H19 overexpression increased the mRNA and protein level of PTEN but suppressed the phosphorylation level of Akt in ASMCs containing miR-21 mimic. The phosphorylated Akt was prominently increased in ASMCs after knocking down PTEN in miR-21 mimic + pcDNA3.1-H19 group. Taken together, H19 could regulate PTEN/Akt pathway via sponging miR-21 to inhibit the proliferation and migration of ASMCs. However, we only exhibited how H19 regulated abnormal pathological change of ASMCs stimulated by PDGF-BB in vitro. Further in vivo study is required and the asthmatic mouse model needs to be established to continuously investigate the effect of H19 on airway remodeling of asthma.
In conclusion, our study elucidated that H19 overexpression could suppress the proliferation and migration ability of ASMCs with PDGF-BB treatment via controlling the miR-21/PTEN/Akt signaling pathway. The present study suggested that H19 might serve as a potential biomarker and a target for interventing hyperplasia of airway smooth muscle cells in children with asthma.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Asher I, Pearce N. Global burden of asthma among children. Int J Tuberc Lung Dis. 2014;18(11):1269–1278. doi:10.5588/ijtld.14.0170
2. Masoli M, Fabian D, Holt S, et al. The global burden of asthma: executive summary of the GINA dissemination committee report. Allergy. 2004;59(5):469–478. doi:10.1111/j.1398-9995.2004.00526.x
3. International Study of Asthma and Allergies in Childhood (ISAAC) Steering Committee. Worldwide variations in the prevalence of asthma symptoms: the international study of asthma and allergies in childhood (ISAAC). Eur Respir J. 1998;12(2):315–335. doi:10.1183/09031936.98.12020315
4. Lambrecht BN, Hammad H. The airway epithelium in asthma. Nat Med. 2012;18(5):684–692. doi:10.1038/nm.2737
5. Munakata M. Airway remodeling and airway smooth muscle in asthma. Allergol Int. 2006;55(3):235–243. doi:10.2332/allergolint.55.235
6. Hirota N, Martin JG. Mechanisms of airway remodeling. Chest. 2013;144(3):1026–1032. doi:10.1378/chest.12-3073
7. James AL, Wenzel S. Clinical relevance of airway remodelling in airway diseases. Eur Respir J. 2007;30(1):134–155. doi:10.1183/09031936.00146905
8. Ito I, Fixman ED, Asai K, et al. Platelet-derived growth factor and transforming growth factor-beta modulate the expression of matrix metalloproteinases and migratory function of human airway smooth muscle cells. Clin Exp Allergy. 2009;39(9):1370–1380. doi:10.1111/j.1365-2222.2009.03293.x
9. Stamatiou R, Paraskeva E, Gourgoulianis K, et al. Cytokines and growth factors promote airway smooth muscle cell proliferation. ISRN Inflamm. 2012;2012:731472. doi:10.5402/2012/731472
10. Govindaraju V, Michoud M-C, Al-Chalabi M, et al. Interleukin-8: novel roles in human airway smooth muscle cell contraction and migration. Am J Physiol Cell Physiol. 2006;291(5):C957–65. doi:10.1152/ajpcell.00451.2005
11. Beermann J, Piccoli M-T, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi:10.1152/physrev.00041.2015
12. Austin PJ, Tsitsiou E, Boardman C, et al. Transcriptional profiling identifies the long noncoding RNA plasmacytoma variant translocation (PVT1) as a novel regulator of the asthmatic phenotype in human airway smooth muscle. J Allergy Clin Immunol. 2017;139(3):780–789. doi:10.1016/j.jaci.2016.06.014
13. Liu Z, Mei L, He Z. Long non-coding RNA00882 contributes to platelet-derived growth factor-induced proliferation of human fetal airway smooth muscle cells by enhancing Wnt/β-catenin signaling via sponging miR-3619-5p. Biochem Biophys Res Commun. 2019;514(1):9–15. doi:10.1016/j.bbrc.2019.04.106
14. Poulet C, Njock MS, Moermans C, et al. Exosomal long non-coding RNAs in lung diseases. Int J Mol Sci. 2020;21(10).
15. Hammad HM. Plasma microRNA-21, microRNA-146a and IL-13 expression in asthmatic children. Innate Immun. 2018;24(3):171–179.
16. Liu F, Qin H-B, Xu B, et al. Profiling of miRNAs in pediatric asthma: upregulation of miRNA-221 and miRNA-485-3p. Mol Med Rep. 2012;6(5):1178–1182. doi:10.3892/mmr.2012.1030
17. Liu Y, Yang K, Shi H, et al. MiR-21 modulates human airway smooth muscle cell proliferation and migration in asthma through regulation of PTEN expression. Exp Lung Res. 2015;41(10):535–545. doi:10.3109/01902148.2015.1090501
18. Wan P, Su W, Zhang Y, et al. LncRNA H19 initiates microglial pyroptosis and neuronal death in retinal ischemia/reperfusion injury. Cell Death Differ. 2020;27(1):176–191. doi:10.1038/s41418-019-0351-4
19. Shao Y, Chong L, Lin P, et al. MicroRNA-133a alleviates airway remodeling in asthma through PI3K/AKT/mTOR signaling pathway by targeting IGF1R. J Cell Physiol. 2019;234(4):4068–4080. doi:10.1002/jcp.27201
20. Liu L, Pan Y, Zhai C, et al. Activation of peroxisome proliferation-activated receptor-γ inhibits transforming growth factor-β1-induced airway smooth muscle cell proliferation by suppressing Smad-miR-21 signaling. J Cell Physiol. 2018;234(1):669–681. doi:10.1002/jcp.26839
21. Fang L, Wang X, Sun Q, et al. IgE downregulates PTEN through MicroRNA-21-5p and stimulates airway smooth muscle cell remodeling. Int J Mol Sci. 2019;20(4):875. doi:10.3390/ijms20040875
22. Holgate ST. Epithelium dysfunction in asthma. J Allergy Clin Immunol. 2007;120(6):1233–1244. doi:10.1016/j.jaci.2007.10.025
23. Liesker JJ, Ten Hacken NH, Zeinstra-Smith M, et al. Reticular basement membrane in asthma and COPD: similar thickness, yet different composition. Int J Chron Obstruct Pulmon Dis. 2009;4:127–135. doi:10.2147/copd.s4639
24. Joubert P, Hamid Q. Role of airway smooth muscle in airway remodeling. J Allergy Clin Immunol. 2005;116(3):713–716. doi:10.1016/j.jaci.2005.05.042
25. Zhang J, Zhu Y, Wang R. Long noncoding RNAs in respiratory diseases. Histol Histopathol. 2018;33(8):747–756. doi:10.14670/HH-11-966
26. Zhang XY, Tang X-Y, Li N, et al. GAS5 promotes airway smooth muscle cell proliferation in asthma via controlling miR-10a/BDNF signaling pathway. Life Sci. 2018;212:93–101. doi:10.1016/j.lfs.2018.09.002
27. Su H, Xu X, Yan C, et al. LncRNA H19 promotes the proliferation of pulmonary artery smooth muscle cells through AT(1)R via sponging let-7b in monocrotaline-induced pulmonary arterial hypertension. Respir Res. 2018;19(1):254. doi:10.1186/s12931-018-0956-z
28. Zhao Y, Feng C, Li Y, et al. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem. 2019;460(1–2):1–8. doi:10.1007/s11010-019-03564-1
29. Pang X, Qiao J. Galectin-1 inhibits PDGF-BB-induced proliferation and migration of airway smooth muscle cells through the inactivation of PI3K/Akt signaling pathway. Biosci Rep. 2020;40(6).
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.