Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA GIHCG Promotes the Development of Esophageal Cancer by Modulating miR-29b-3p/ANO1 Axis
Authors Zhao W, Huang Z, Liu H, Wang C
Received 16 September 2020
Accepted for publication 27 November 2020
Published 31 December 2020 Volume 2020:13 Pages 13387—13400
DOI https://doi.org/10.2147/OTT.S282348
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Weifeng Zhao,1 Zhoufeng Huang,1,2 Huimin Liu,1 Chaojie Wang1
1Department of Oncology, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University, Zhengzhou City, Henan Province 450003, People’s Republic of China; 2Institute of Hematology, Henan Provincial People’s Hospital, Zhengzhou City, Henan Province 450003, People’s Republic of China
Correspondence: Chaojie Wang
Department of Oncology, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University, No. 7 Weiwu Road, Jinshui District, Zhengzhou City, Henan Province 450003, People’s Republic of China
Email [email protected]
Background: Esophageal cancer is one of the most frequent cancers with a higher mortality worldwide. Although many long non-coding RNAs (LncRNAs) are reported to play important roles in the progression of esophageal cancer, the function of lncRNA GIHCG in esophageal cancer remains unclear.
Methods: The expression of GIHCG in esophageal cancer tissues and cancer cell lines was detected by qRT-PCR. Cell proliferation was evaluated by Cell Counting Kit-8 (CCK-8) assay, EdU staining assay and colony formation assay. Cell invasion and migration were measured by transwell assay. Cell apoptosis was detected by a flow cytometer. Luciferase reporter assay and RIP assay were used to determine the interaction between GIHCG and miR-29b-3p, and their subsequent regulation of anoctamin 1 (ANO1). The expression of ANO1 in esophageal cancer tissues and cell lines was detected by Western blot. The effect of GIHCG/miR-29b-3p in tumor formation was assessed by the xenograft nude mice model in vivo.
Results: GIHCG was significantly upregulated in esophageal cancer tissues and relevant cancer cell lines. Downregulation of GIHCG significantly inhibited the growth, colony formation, invasion, migration and induced apoptosis of esophageal cancer cells in vitro. Bioinformatic analysis and RIP assay determined that GIHCG was a sponge of miR-29b-3p, and ANO1 was a direct target of miR-29b-3p. Moreover, functional experiments showed that GIHCG upregulated ANO1 expression by directly sponging miR-29b-3p. Furthermore, in vivo experiment revealed that knockdown of GIHCG significantly inhibited tumor growth in nude mice.
Conclusion: Our study revealed that lncRNA GIHCG promoted the progression of esophageal cancer by targeting the miR-29b-3p/ANO1 axis, suggesting that GIHCG might be a novel therapeutic target for esophageal cancer.
Keywords: esophageal cancer, GIHCG, miR-29b-3p, ANO1, tumorigenesis
Introduction
Esophageal cancer has become the sixth leading cause of death-related death and the eighth most common cancer worldwide.1 Recently, medical oncologists and radiation oncologists have been increasingly involved due to the implementation of neo-adjuvant therapy, which has been shown to efficiently improve the overall survival of patients with esophageal cancer.2 Despite many advantages in the management and treatment, the 5-year survival rates of patients with esophageal cancer are still poor with only approximately 10%.3,4 Hence, it is more urgent to understand the underlying molecular mechanisms involved in esophageal cancer development, which may contribute to identify new targets for treating esophageal cancer.
Long-chain non-coding RNAs (lncRNAs) are a class of independent transcripts with more than 200 nucleotides in length and lack protein coding function.5 Recently, lncRNAs have been identified to be significantly associated with tumor behaviors and prognosis.6–8 Gradually increased during hepatocarcinogenesis (GIHCG) is a newly identified lncRNA, and increasing studies demonstrated that it can promote the progression of various human cancers. For example, GIHCG is highly expressed in ovarian cancer tissues compared with normal tissues, and downregulation of GIHCG can efficiently inhibit the progression of ovarian cancer through stimulating cell cycle and cell proliferation by targeting miR-429.9 GIHCG promotes cell proliferation and migration of gastric cancer cells by directly modulating miR-1281.10 GIHCG has been reported to enhance cell cycle, proliferation and migration of tongue squamous cell carcinoma cells through directly targeting miR-429 in vitro.11 In addition, GIHCG has also been found to play crucial oncogenic roles in several other malignancies such as breast cancer,12 hepatocellular carcinoma,13 colorectal cancer14 and renal cell carcinoma.15 Although GIHCG exhibits important roles in a series of human cancers, its role in esophageal cancer remains unclear.
MicroRNAs (miRNAs) are a group of widely known non-coding RNA with 22 nucleotides in length, and have been identified to play essential regulatory roles in the regulation of post-transcriptional gene expression.16 Increasing reports have revealed that miRNAs can participate in multiple biological processes during cancer development including cell growth, metastasis, invasion and migration.17 MiR-29b-3p is shown to be significantly involved in the progression of human tumors. For example, upregulation of miR-29b-3p induces chondrocyte apoptosis and promotes the progression of osteoarthritis through targeting PGRN.18 Downregulation of miR-29b-3p significantly inhibits the development of triple-negative breast cancer by directly targeting TRAF3.19 MiR-29b-3p is markedly overexpressed in plasma-derived extracellular vesicles of prostate cancer patients compared with that of healthy subjects, which may be considered as a potential biomarker for prostate cancer.20
Anoctamin 1 (ANO1), also named as TMEM16A, ORAOV2, TAOS2, DOG1 or FLJ10261, is located on human chromosome 11q13, which contains 26 exons and encode a 960 amino acid protein.21 ANO1 is related to calcium-dependent chloride channel activity and can participate in multiple biological processes including cell proliferation, motility and attachment.22,23 ANO1 has been found to be highly expressed in tumor tissues of esophageal carcinomas such as esophageal squamous cell carcinoma,24 head and neck squamous cell carcinoma25 and squamous cell carcinoma,26 and is considered as a potential biomarker for prognosis of esophageal carcinomas. Moreover, overexpression of ANO1 has been demonstrated to be associated with the occurrence, proliferation and migration of several tumor cells including esophageal cancer cells.27 However, the upstream regulatory axis of ANO1 in esophageal cancer has not been well studied.
In the present study, we firstly found that lncRNA GIHCG was significantly upregulated in esophageal cancer tissues and cancer cell lines. Moreover, our results demonstrated that downregulation of GIHCG inhibited the proliferation, invasion, migration while induced apoptosis of esophageal cancer cells in vitro, and also suppressed tumor growth in vivo by directly targeting miR-29b-3p/ANO1 axis, suggesting that GIHCG/miR-29b-3p axis might be potential therapeutic targets for esophageal cancer.
Materials and Methods
Tissue Specimens
A total of 45 patients with esophageal cancer enrolled at Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University, from 2017 to 2019 were recruited in this study. The clinicopathological features of esophageal cancer patients are shown in supplementary Table 1. All participants had given their written informed consent. All tumor tissues and paired adjacent normal tissues were removed from bodies by surgical excision and then immediately snap-frozen in liquid nitrogen before storage at –80 °C. Meanwhile, all tissue samples were histologically confirmed by two pathologists. All paired tissue samples were used to compare the expression levels of GIHCG, miR-29b-3p and ANO1. In addition, the clinical significance and prognostic value of GIHCG was evaluated by Kaplan–Meier survival analysis as previously described.28 This study was approved by the Ethics Committee of Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University.
Cell Culture
Human esophageal cancer cell lines (Eca-109, Ec-9706, EC8712, TE-13 and TE-10) and normal esophageal epithelial cell line HEEC were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM medium (Invitrogen) supplementing with 10% FBS (Invitrogen) at 37°C with 5% CO2.
Cell Transfection
Ec-9706 and TE-10 cells were transfected with 50nM small interfering RNA targeting GIHCG (si-GIHCG), miR-29b-3p mimics, miR-29b-3p inhibitor and respective negative controls (si-NC, miR-NC and inhibitor NC) at the final concentration of 100 nM transfection reagent by the Lipofectamine 2000 kit (Invitrogen) according to the manufacturer’s instructions. The sequence of si-GIHCG and si-NC were as follows: si-GIHCG: 5′-GCATCCCGCGTCAATCTGAAGGAACCTCAAGGGA-3′; si-NC: 5′-GTTCCATCAGGATGACGCCCCTTTTGGGAAAGCCT3′. MiR-29-3p mimics, miR-NC, miR-29-3p inhibitor, and inhibitor NC were purchased from GenePharma (Suzhou, China). To construct GIHCG and ANO1 overexpressing vector, the cDNA sequences of GIHCG and ANO1 were amplified with human genome as the template and cloned into pcDNA3.1 expression vector (Geneseed Biotech, Guangzhou), and empty vector pcDNA3.1 was considered as the negative control. Plasmids were transfected into Ec-9706 and TE-10 cells also by Lipofectamine 2000 kit (Invitrogen). Cells were harvested for analyses 48 h after transfections.
qRT-PCR Analysis
Total RNA of tissues or cultured cells was extracted by using TRIzol reagent (Invitrogen). Total RNA was reversely transcribed to cDNA by the Primescript RT Reagent (Takara Bio, Inc.), and qRT–PCR analysis was performed using SYBR® Green Real‐time PCR Master Mix (Toyobo Co. Ltd., Osaka, Japan) on a 7500 Fast Real-Time PCR System (Applied Biosystems). The relative fold expression change of target genes was analyzed by using the 2−ΔΔCt method, with GAPDH and U6 as the internal reference.29 The primers used in this study were as follows: GIHCG forward: 5′- CTTTATCACCTGCCGGGAA-3′, reverse: 5′-CGGTAAGAATCCAAAGG-3′; miR-29b-3p forward: 5′-CTGAATGTGGAGAGAATGT-3′, reverse: 5′-GTTCCTCGACATTCGGGCCG-3′; ANO1 forward: 5′-CCACAGGATTCGGGGAACT-3′, reverse: 5′-GAAGTCATTACAGCGGTGCC-3′; GAPDH forward: 5′-CGAGAGAATCCGCGGACAT-3′, reverse: 5′-TTGTGCAATACAGCGTGGAC-3′; U6 forward: 5′-GACAGATTCGGTCTGTGGCAC-3′, reverse: 5′-GATTACCCGTCGGCCATCGATC-3′.
Western Blot
Total protein of tissues or cultured cells was extracted by RIPA Lysis Buffer containing protease inhibitor PMSF (Thermo, USA). Subsequently, approximately equal amounts of protein were separated by 10% SDS-PAGE and transferred into PVDF membranes. After blocking with 5% non-fat milk for 5 min, the membranes were then incubated with primary antibody ANO1 (1:5000, AF6207, Beytotime) and GAPDH (1:1000, SG2267, RealJimes) at 4°C overnight. Of which, GAPDH was regarded as the internal reference. On the next day, the membranes were incubated with an appropriate HRP‐conjugated secondary antibody for 1 h at room temperature. Finally, the bonds were visualized by using the ECL kits (Amersham), and the optical density of the protein bands was quantified under the ImageJ software.
CCK-8 Assay
Cell proliferation was evaluated by using Cell Counting Kit-8 (Dojindo, Beijing, China) according to the manufacturer’s instructions as previously described.18 Approximately, a certain number of transfected cells were seeded into 96-well plates and maintained in DMEM medium containing 10% FBS. Ten microliters of CCK‐8 solution was added to each well at the time point of 24, 48, 72 and 96h, then the absorbance was detected by EnSpire Multimode Plate Reader (PerkinElmer, Waltham, Massachusetts, USA) at 450 nm.
Colony Formation Assay
Briefly, approximately 1000 cells were seeded into a 6-well plate and cultured for 2 weeks. Subsequently, cells were fixed with 4% paraformaldehyde for 15 min and stained with 1% crystal violate for 10–30 min. The images of colony-forming units were captured by using a light microscope and the number of colonies was counted in three random wells.
Edu Staining Assay
The EdU (5-ethynyl-2′-deoxyuridine) proliferation assay was performed by using Cell-Light EdU Apollo 567 In Vitro Imaging Kit (Ribobio) as previously described.30 Briefly, 1×105 cells were seeded into 96-well plates and cultured for 24 h. Edu solution was added into each well with the final concentration at 50 μM and incubated for 2 h. Then, 4% paraformaldehyde fixing solution was added for 20 min, 0.5% Triton X-100 was added for 15 min. Finally, 100 μL of Apollo reaction solution was added to each well and incubated for 30 min, then Hoechst33342 staining solution (Beyotime, Shanghai, China) was added for another 30 min. Images were captured using fluorescence microscopy (Nikon) and merged using Adobe Photoshop 6.0 software. Afterwards EdU-positive cells and total cells were counted within each field.
Transwell Assay
Cell invasion and migration was evaluated by using transwell assay as previously described.31 And, 3×105 Ec-9706 and TE-10 cells transfected with si-GIHCG, miR-29p-3p mimics, miR-29p-3p inhibitor and corresponding negative controls were transferred to the upper Matrigel-coated or not Matrigel-coated invasion chambers (BD Biosciences, San Jose, CA, USA) in a serum-free DMEM medium, and DMEM medium containing 10% FBS was added to the lower chambers. After 24 h, non-migrated or non-invaded cells on the upper surface were removed, and the migrating or invading cells on the underside surface were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and then imaged in 10 random fields under a microscope.
Apoptotic Analysis
Cell apoptosis was analyzed by using Annexin V-FITC/PI apoptosis detection kit (Keygen Biot Solasodine, Nanjing, China) according to the manufacturer’s instructions. In brief, 1 × 106 transfected Ec-9706 and TE-10 cells were plated into 6 well plates and cultured for 24 h. After trypsinization, cell suspension was added with 5 μL Annexin V-FITC followed by 5 μL PI solution in dark and incubated for 20 min. Flow cytometric analysis for apoptosis rate was performed by using a FACSCalibur flow cytometer within 1 h.
Luciferase Reporter Assay
The wild type (WT) and mutant (MUT) of GIHCG or ANO1 containing the putative binding site with miR-29b-3p were amplified by PCR and cloned into pmirGLO luciferase reporter vector (Promega Corporation, Madison, WI, USA). Then, the luciferase reporter plasmids were co-transfected with miR-29b-3p mimics or miR-NC into Ec-9706 and TE-10 cells by using Lipofectamine 2000 kit (Invitrogen). Following 48 h transfection, relative luciferase activity was determined using the Dual-Luciferase Reporter assay system and normalized to renilla luciferase activity, respectively.
The Construction of Stably Transfected Cell Line
To generate the stable knockdown of GIHCG in cell line Ec-9706, the lentivirus vectors expressing short hairpin RNA (shRNA) targeting GIHCG (sh-GIHCG) or scrambled oligonucleotides (sh-NC) were constructed and purchased from GenePharma (Shanghai, China). Lentivirus were transfected into Ec-9706 cells by using Lipofectamine 2000 kit (Invitrogen). In brief, cells were seeded in six-well plates, then transfected with 1×108 transducing units (TUs)/mL lentivirus (10 μL). At 48h after transfection, cells were treated with puromycin (2 μg/mL) for 2 weeks to select for stable cell line. The sequence of sh-GIHCG was as follows:sh-GIHCG: sense: 5′-GGATCGGGTTAGGAGCCTAGGATGCCTTCCAGGGA-3′, anti-sense: 5′-ACCATCCTTAGCTCCTGGATTTTTGGGGAAAGCCT3′; To construct the Ec-9706 cells stably transfected miR-29b-3p inhibitor, Ec-9706 cells were transfected with 50 nM miR-29b-3p inhibitor by using Lipofectamine 2000 kit and selected with 2 μg/mL puromycin for 2 weeks. To construct concurrently stably knockdown of both miR-29b-3p and GIHCG in Ec-9706 cells, GIHCG stable knockdown Ec-9706 cells were transfected with 50 nM miR-29b-3p inhibitor by using Lipofectamine 2000 kit and selected with 2 μg/mL puromycin for 2 weeks.
Xenograft Tumor Model in Mice
A total of 12 BALB/c nude mice (female, 4–6 weeks and approximately 16–22g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The xenograft model with esophageal cancer was established as previously described.32 Briefly, approximately 5 × 106 Ec-9706 cells stably transfected with sh-GIHCG, miR-29b-3p inhibitor, sh-NC, or co-transfected with sh-GIHCG and miR-29b-3p inhibitor were subcutaneously inoculated into the nude mice, and generated four groups (n = 3): sh-GIHCG group, miR-29b-3p inhibitor group, sh-NC group, sh-GIHCG + miR-29b-3p inhibitor group. Tumor volume of different groups was evaluated once a week for 5 weeks following the formula: V = 0.52 × L × W2 (L is the length of tumor and W is the tumor weight). After 5 weeks, mice were euthanized by cervical dislocation, and tumors of different groups were taken out, then tumor weight was evaluated. Finally, tumors were paraffin-embedded, cut into 4-μm thickness, and incubated with monoclonal antibodies against human Ki-67 (1:200, Abcam) to perform the Ki-67 staining assay as previously described.33 This study was approved by the Animal Ethics Committee of Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University. All animal studies were performed according to the Institutional Animal Care and Use Committee of Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of People’s Hospital of Henan University and approved by the Animal Ethics Committee of People’s Hospital of Henan University.
Statistical Analysis
Data were analyzed by SPSS19.0 statistical software and presented as mean ± SD method. All experiments have three replicates. Difference was determined by Student’s t-test (two groups) or two-way ANOVA (multiple groups). P value <0.05 was considered as significant threshold. Kaplan–Meier method was applied to perform the survival analysis of patients with esophageal cancer.
Results
LncRNA GIHCG Was Significantly Upregulated in Esophageal Cancer Tissues
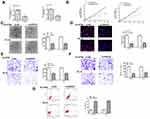
To explore the role of GIHCG in esophageal cancer, we firstly detected the expression of GIHCG in esophageal cancer tissues by qRT-PCR and the results showed that GIHCG was highly overexpressed in tumor tissues compared with matched adjacent normal tissues (n = 45) (p < 0.01) (Figure 1A). Meanwhile, the expression level of GIHCG in tumor tissues and matched normal tissues was analyzed, and the results indicated that the levels of GIHCG were almost all higher in tumor tissues than that in matched adjacent normal tissues (n = 45) (p < 0.01) (Figure 1B). Then, the expression of GIHCG in cancer cell lines was also detected and the results showed that GIHCG was also markedly upregulated in cancer cell lines Eca-109 (p < 0.01), Ec-9706 (p < 0.001), EC8712 (p < 0.01), TE-13 (p < 0.01) and TE-10 cells (p < 0.001) compared with normal esophageal epithelial cell line HEEC (Figure 1C), and exhibited a highest expression in Ec-9706 and TE-10 cells. Then, Ec-9706 and TE-10 cells were selected for the subsequent experiments. In addition, Kaplan–Meier survival curve indicated that esophageal cancer patients with high GIHCG level had a poorer prognosis compared with that with low GIHCG level (p < 0.01) (Figure 1D). These results suggested that GIHCG might play a potential oncogenic role in esophageal cancer.
Downregulation of GIHCG Inhibited the Growth of Esophageal Cancer Cells in vitro
To further confirm the oncogenic role in esophageal cancer, si-GIHCG (small interfering RNA targeting GIHCG) or si-NC (negative control) was transfected into Ec-9706 and TE-10 cells, and qRT-PCR indicated that si-GIHCG significantly decreased the expression of GIHCG compared with si-NC in both Ec-9706 (p < 0.001) and TE-10 cells (p < 0.001) (Figure 2A). Downregulation of GIHCG obviously inhibited cell viability of Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with si-NC (Figure 2B). Si-GIHCG significantly decreased colony number of both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with si-NC (Figure 2C). Meanwhile, si-GIHCG significantly decreased the EdU-positive cells of both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with si-NC (Figure 2D). Transwell assay revealed that si-GIHCG not only inhibited the invasion of both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with si-NC (Figure 2E) but also inhibited the migration of both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with si-NC (Figure 2F). In addition, downregulation of GIHCG significantly enhanced the apoptosis of both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with si-NC (Figure 2G). These results indicated that downregulation of GIHCG inhibited the growth of esophageal cancer cells in vitro.
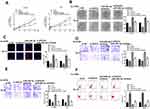
GIHCG Served as a Sponge of miR-29b-3p
To explore the mechanism of GIHCG in esophageal cancer, Starbase was used to predict the potential targets of GIHCH and the results showed that there was a putative binding site between GIHCG and miR-29b-3p (Figure 3A), suggesting that miR-29b-3p might be a potential target of GIHCG. Then, Ec-9706 and TE-10 cells were transfected with miR-29b-3p mimics or miR-NC, and qRT-PCR revealed that miR-29b-3p mimics significantly increased the expression of miR-29b-3p in both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with miR-NC (Figure 3B). To determine their relationship, luciferase reporter assay was performed and the results indicated that miR-29b-3p mimics significantly decreased the relative luciferase activity of WT GIHCG vector in both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01), while exhibited no obvious change on MUT GIHCG vector in two cell lines (Figure 3C). Meanwhile, si-GIHCG significantly increased the expression of miR-29b-3p in both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with si-NC, and overexpression of GIHCG (pc-GIHCG) markedly decreased miR-29b-3p in both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with pc-NC (empty vector control) (Figure 3D). In addition, miR-29b-3p expression was markedly downregulated in cancer cell lines including Eca-109 (p < 0.01), Ec-9706 (p < 0.001), EC8712 (p < 0.01), TE-13 (p < 0.01) and TE-10 cells (p < 0.001) compared with that in HEEC cells (Figure 3E). Moreover, miR-29b-3p was markedly downregulated in tumor tissues compared with that in adjacent normal tissues (n = 45) (p < 0.01) (Figure 3F). Pearson’s correlation analysis showed that there was an obviously negative correlation between GIHCG and miR-29b-3p expression in esophageal cancer tissues (n = 45) (p < 0.01) (Figure 3G). These results revealed that GIHCG was a sponge of miR-29b-3p and the effect of GIHCG in esophageal cancer might be partially mediated by miR-29b-3p.
Downregulation of miR-29b-3p Reversed Si-GIHCG Induced Inhibitory Effect in Esophageal Cancer Progression in vitro
To explore whether the effect of GIHCG was mediated by miR-29b-3p, Ec-9706 and TE-10 cells were transfected with si-GIHCG, si-NC, miR-29b-3p inhibitor, or co-transfected with si-GIHCG and miR-29b-3p inhibitor. The results of CCK-8 assay (Figure 4A), colony formation assay (Figure 4B) and EdU staining assay (Figure 4C) revealed that miR-29b-3p inhibitor significantly exacerbated the growth of Ec-9706 and TE-10 cells (p < 0.01), while co-transfection of si-GIHCG and miR-29b-3p inhibitor obviously attenuated the inhibitory effect of si-GIHCG on cell proliferation (p < 0.01). MiR-29b-3p inhibitor markedly promoted the invasion and migration capacity of Ec-9706 and TE-10 cells (p < 0.01), while co-transfection of si-GIHCG and miR-29b-3p inhibitor obviously attenuated the inhibitory effect of si-GIHCG on cell invasion and migration (p < 0.01) (Figure 4D and E). Meanwhile, miR-29b-3p inhibitor significantly decreased apoptosis rate of Ec-9706 and TE-10 cells (p < 0.05), while co-transfection of si-GIHCG and miR-29b-3p inhibitor obviously reversed the effect of si-GIHCG on apoptosis (p < 0.05) (Figure 4F). These results suggested that the effect of GIHCG in esophageal cancer might be partially mediated by miR-29b-3p.
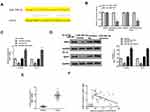
ANO1 Was Target of miR-29b-3p
Next, TargetScan was used to predict the potential targets of miR-29b-3p, and the results showed that ANO1 might be a target of miR-29b-3p (Figure 5A). Then, luciferase reporter assay was performed and indicated that miR-29b-3p mimics significantly decreased the relative luciferase activity of WT ANO1 vector in both Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with the miR-NC group, while exhibited no obvious change in MUT ANO1 vector in two cell lines (Figure 5B). Meanwhile, miR-29b-3p mimics significantly reduced the expression of ANO1 at both mRNA level and protein level in Ec-9706 and TE-10 cells compared with miR-NC (Figure 5C and D), while miR-29b-3p inhibitor increased the expression of ANO1 at both mRNA level and protein level in two cell lines compared with miR-NC (Figure 5C and D). In addition, the expression of ANO1 was significantly upregulated in tumor tissues compared with adjacent normal tissues (n = 45) (p < 0.01) (Figure 5E). Pearson’s correlation analysis indicated the there was also an obviously negative correlation between miR-29b-3p and ANO1 expression in esophageal cancer tissues (n = 45) (p < 0.01) (Figure 5F). These results suggested that ANO1 was the target of miR-29b-3p and the effect of miR-29b-3p in esophageal cancer might be mediated by ANO1.
Overexpression of ANO1 Reversed miR-29b-3p Mimics Induced Inhibitory Effect in Esophageal Cancer Progression in vitro
To further determine the effect of miR-29b-3p in esophageal cancer was mediated by ANO1, Ec-9706 and TE-10 cells were transfected with miR-29b-3p mimics, miR-NC, or co-transfected with miR-29b-3p mimics and pcDNA-ANO1 (overexpression of ANO1). The results of CCK-8 assay (Figure 6A), colony formation assay (Figure 6B) and EdU staining assay (Figure 6C) showed that miR-29b-3p mimics significantly inhibited the proliferation of both Ec-9706 and TE-10 cells compared with miR-NC (p < 0.01), while co-transfection of miR-29b-3p mimics and pcDNA-ANO1 obviously attenuated miR-29b-3p mimics induced inhibitory effect on cell proliferation (p < 0.05). Transwell assay indicated that miR-29b-3p mimics not only markedly decreased the invasion capacity (Figure 6D) but also decreased the migration capacity of Ec-9706 and TE-10 cells (Figure 6E), while co-transfection of miR-29b-3p mimics and pcDNA-ANO1 obviously reversed miR-29b-3p mimics induced inhibitory effect on invasion and migration (p < 0.05) (Figure 6D and E). Meanwhile, miR-29b-3p mimics significantly promoted the apoptosis of Ec-9706 (p < 0.01) and TE-10 cells (p < 0.01) compared with miR-NC, while co-transfection of miR-29b-3p mimics and pcDNA-ANO1 obviously reversed the effect of miR-29b-3p mimics on apoptosis (p < 0.05) (Figure 6F). These results revealed that overexpression of ANO1 efficiently reversed miR-29b-3p mimics induced inhibitory effect in esophageal cancer progression in vitro.
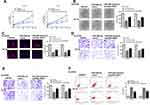
GIHCG/miR-29b-3p Modulated Tumor Growth in vivo
Finally, to confirm the role of GIHCG in esophageal cancer, a xenograft tumor model in vivo was established through subcutaneously injected with Ec-9706 cells stably transfected with sh-GIHCG, sh-NC, miR-29b-3p inhibitor, or co-transfected with sh-GIHCG and miR-29b-3p inhibitor. After 5 weeks, the tumors of different groups were removed and the representative images are shown in Figure 7A. Compared with the sh-NC group, tumor weight in the sh-GIHCG group was significantly reduced (p < 0.01), and miR-29b-3p inhibitor increased tumor weight (p < 0.01), while co-transfection of sh-GIHCG and miR-29b-3p inhibitor obviously reversed sh-GIHCG induced inhibitory effect (p < 0.05) (Figure 7B). Similarly, sh-GIHCG decreased tumor volume compared with sh-NC group (p < 0.01), and miR-29b-3p inhibitor increased the tumor volume (p < 0.01), while co-transfection of sh-GIHCG and miR-29b-3p inhibitor obviously reversed sh-GIHCG induced inhibitory effect (p < 0.05) (Figure 7C). Meanwhile, Ki-67 staining assay in tumor tissues showed that sh-GIHCG obviously decreased the number of Ki-67 positive cells compared with sh-NC, and miR-29b-3p inhibitor increased Ki-67-positive cells, while co-transfection of sh-GIHCG and miR-29b-3p inhibitor obviously reversed sh-GIHCG induced inhibitory effect on proliferation in vivo (Figure 7D). In addition, the expression of ANO1 in tumor tissues was also detected by Western blot, and the results indicated that sh-GIHCG significantly decreased the protein level of ANO1 compared with si-NC (p < 0.01), and miR-29b-3p inhibitor increased ANO1 expression (p < 0.05), while co-transfection of sh-GIHCG and miR-29b-3p inhibitor obviously reversed sh-GIHCG induced inhibitory effect on ANO1 expression (p < 0.05) (Figure 7E). These results revealed that knockdown of GIHCG inhibited tumor growth through modulating miR-29b-3p in vivo.
Discussion
Esophageal cancer has become one of the most aggressive cancer types worldwide,34 and the etiology remains poorly understood. Previous studies have revealed that a large number of lncRNAs are closely associated with diverse cellular processes including proliferation, invasion, metastasis and so on in human cancers.35 In this study, our results revealed that GIHCG was significantly upregulated in esophageal cancer tissues and cancer cell lines. Moreover, knockdown of GIHCG markedly inhibited cell proliferation, invasion, migration and induced apoptosis of esophageal cancer cells in vitro, and also inhibited tumor growth in vivo, suggesting an oncogenic role of GIHCG in esophageal cancer.
In esophageal cancer, the dysregulation of many lncRNAs is often observed and some of which have been well studied. For instance, SNHG7 promotes the proliferation and suppresses cell apoptosis of esophageal cancer cells via modulating the expression of p15 and p16 in vitro.36 H19 promotes epithelial-mesenchymal transition (EMT) and metastasis of esophageal cancer cells by targeting the STAT3/EZH2 axis.37 CASC9 has been revealed that can exacerbate the metastasis and EMT process through upregulating LAMC2 by directly interacting with the CREB-binding protein.38 MALAT1 promotes the EMT process of esophageal cancer cells by regulating the Ezh2-Notch1 signaling pathway.39 Su et al revealed that downregulation of MIR22HG could efficiently inhibit the proliferation and induce apoptosis of esophageal cancer cells by activating the STAT3/c-Myc/FAK signaling pathway.40 CCAT1 promotes the drug resistance of esophageal cancer cells through targeting the miR-143/PLK1/BUBR1 axis, and CCAT1 may be considered as a potential biomarker for the proliferation of esophageal cancer cells.41 PVT1 has emerged as an newly identified oncogene in many tumor types, and Xu et al found that upregulation of PVT1 predicted a poor prognosis and suggested that PVT1 might be a potential therapeutic target for esophageal cancer.42 In addition, lncRNAs have been identified as survival predictor and may be novel prognostic signatures in esophageal cancer.43,44 Although lncRNA GIHCG has been reported that can participate in tumor progression of various types of human cancers, its role and underlying molecular mechanisms in esophageal cancer remain unclear.
It has been reported that lncRNAs always serve as ceRNAs of miRNAs to play their essential roles in the pathogenesis of cancers.45 LncRNA MT1JP functions as a ceRNA to regulate FBXW7 expression by competitively binding to miR-92a-3p in gastric cancer.46 LncRNA TUG1 influences cell proliferation, migration and EMT process of papillary thyroid cancer cells by directly targeting miR-145.47 To further explore the specific mechanism of GIHCG in esophageal cancer, Starbase was used to predict its targets. The results of prediction showed that miR-29b-3p might be a direct target of GIHCG, and luciferase reporter assay determined that miR-29b-3p mimics significantly decreased the relative luciferase activity of WT GIHCG vector in both Ec-9706 and TE-10 cells, while exhibited no obvious change in MUT GIHCG vector in two cell lines. Meanwhile, downregulation of GIHCG significantly increased the expression of miR-29b-3p in both Ec-9706 and TE-10 cells compared with si-NC, and overexpression of GIHCG markedly decreased miR-29b-3p in two cell lines. Moreover, downregulation of miR-29b-3p could efficiently reverse si-GIHCG induced inhibitory effect in esophageal cancer progression including proliferation, invasion, migration and apoptosis in vitro. These results suggested that the effect of GIHCG in esophageal cancer was partially mediated by miR-29b-3p.
Increasing evidences have reported that miRNAs generally regulate the expression of targets to affect the development of human cancers by binding to the 3UTR of targeted mRNAs.48,49 ANO1 has been identified to act as direct targets of multiple miRNAs to participate in the biological process involved in cancer progression. For example, miR‑144 inhibits aggressive phenotypes of tumor cells through directly targeting ANO1 in colorectal cancer.50 Park et al revealed that knockdown of miR-9 promoted the EMT process by targeting ANO1 in colorectal cancer.51 MiR-381 suppresses the metastasis of gastric cancer cells through targeting ANO1.52 Here, our results identified that ANO1 was a target of miR-19b-3p, and overexpression of ANO1 significantly reversed miR-29b-3p mimics induced inhibitory effect in esophageal cancer progression in vitro. Moreover, to confirm the oncogenic role of GIHCG in esophageal cancer, a xenograft tumor model was constructed and the results indicated that downregulation of GIHCG significantly inhibited tumor growth through modulating miR-29b-3p in vivo.
However, overexpression of ANO1 whether reversed the inhibitory effect of sh-GIHCG and miR-29b-3p mimics in tumor growth in vivo should be determined in the subsequent experiments.
Conclusion
In summary, our study provided a new mechanism of lncRNA GIHCG in esophageal cancer: GIHCG promoted the progression of esophageal cancer through upregulating ANO1 expression by directly sponging miR-29b-3p, suggesting that GIHCG might be a novel therapeutic target for esophageal cancer.
Disclosure
The authors declare that they have no competing interests.
References
1. Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21(26):7933–7943. doi:10.3748/wjg.v21.i26.7933
2. Borggreve AS, Kingma BF, Domrachev SA, et al. Surgical treatment of esophageal cancer in the era of multimodality management. Ann N Y Acad Sci. 2018;1434(1):192–209. doi:10.1111/nyas.13677
3. Hoeben A, Polak J, Van De Voorde L, Hoebers F, Grabsch HI, de Vos-geelen J. Cervical esophageal cancer: a gap in cancer knowledge. Ann Oncol. 2016;27(9):1664–1674. doi:10.1093/annonc/mdw183
4. Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412. doi:10.1016/S0140-6736(12)60643-6
5. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi:10.1016/j.cell.2013.06.020
6. Nie L, Wu HJ, Hsu JM, et al. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4(2):127–150.
7. Yang P, Xu ZP, Chen T, He ZY. Long noncoding RNA expression profile analysis of colorectal cancer and metastatic lymph node based on microarray data. Onco Targets Ther. 2016;9:2465–2478. doi:10.2147/OTT.S102348
8. Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi:10.1016/j.ccell.2015.02.004
9. Yao N, Yu L, Zhu B, Gan HY, Guo BQ. LncRNA GIHCG promotes development of ovarian cancer by regulating microRNA-429. Eur Rev Med Pharmacol Sci. 2018;22(23):8127–8134. doi:10.26355/eurrev_201812_16504
10. Liu G, Jiang Z, Qiao M, Wang F. Lnc-GIHCG promotes cell proliferation and migration in gastric cancer through miR- 1281 adsorption. Mol Genet Genomic Med. 2019;7(6):e711. doi:10.1002/mgg3.711
11. Ma L, Wang Q, Gong Z, Xue L, Zuo Z. Long noncoding RNA GIHCG enhanced tongue squamous cell carcinoma progression through regulating miR-429. J Cell Biochem. 2018;119(11):9064–9071. doi:10.1002/jcb.27164
12. Fan LY, Shi KY, Xu D, et al. LncRNA GIHCG regulates microRNA-1281 and promotes malignant progression of breast cancer. Eur Rev Med Pharmacol Sci. 2019;23(24):10842–10850. doi:10.26355/eurrev_201912_19788
13. Sui CJ, Zhou YM, Shen WF, et al. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J Mol Med (Berl). 2016;94(11):1281–1296. doi:10.1007/s00109-016-1442-z
14. Jiang X, Li Q, Zhang S, Song C, Zheng P. Long noncoding RNA GIHCG induces cancer progression and chemoresistance and indicates poor prognosis in colorectal cancer. Onco Targets Ther. 2019;12:1059–1070. doi:10.2147/OTT.S192290
15. He ZH, Qin XH, Zhang XL, Yi JW, Han JY. Long noncoding RNA GIHCG is a potential diagnostic and prognostic biomarker and therapeutic target for renal cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(1):46–54. doi:10.26355/eurrev_201801_14099
16. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi:10.1016/j.molmed.2014.06.005
17. Li C, Lei B, Huang S, et al. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am J Transl Res. 2015;7(10):1747–1764.
18. Chen L, Li Q, Wang J. MiR-29b-3p promotes chondrocyte apoptosis and facilitates the occurrence and development of osteoarthritis by targeting PGRN. J Cell Mol Med. 2017;21(12):3347–3359. doi:10.1111/jcmm.13237
19. Zhang B, Shetti D, Fan C, Wei K. miR-29b-3p promotes progression of MDA-MB-231 triple-negative breast cancer cells through downregulating TRAF3. Biol Res. 2019;52(1):38. doi:10.1186/s40659-019-0245-4
20. Worst TS, Previti C, Nitschke K, et al. miR-10a-5p and miR-29b-3p as extracellular vesicle-associated prostate cancer detection markers. Cancers. 2019;12(1):43. doi:10.3390/cancers12010043
21. Takayama Y, Derouiche S, Maruyama K, Tominaga M. Emerging perspectives on pain management by modulation of TRP channels and ANO1. Int J Mol Sci. 2019;20(14):3411. doi:10.3390/ijms20143411
22. Ayoub C, Wasylyk C, Li Y, et al. ANO1 amplification and expression in HNSCC with a high propensity for future distant metastasis and its functions in HNSCC cell lines. Br J Cancer. 2010;103(5):715–726. doi:10.1038/sj.bjc.6605823
23. Liu W, Lu M, Liu B, Huang Y, Wang K. Inhibition of Ca(2+)-activated Cl(-) channel ANO1/TMEM16A expression suppresses tumor growth and invasiveness in human prostate carcinoma. Cancer Lett. 2012;326(1):41–51. doi:10.1016/j.canlet.2012.07.015
24. Kashyap MK, Marimuthu A, Kishore CJ, et al. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol Ther. 2009;8(1):36–46. doi:10.4161/cbt.8.1.7090
25. Ruiz C, Martins JR, Rudin F, et al. Enhanced expression of ANO1 in head and neck squamous cell carcinoma causes cell migration and correlates with poor prognosis. PLoS One. 2012;7(8):e43265. doi:10.1371/journal.pone.0043265
26. Shi ZZ, Shang L, Jiang YY, et al. Consistent and differential genetic aberrations between esophageal dysplasia and squamous cell carcinoma detected by array comparative genomic hybridization. Clin Cancer Res. 2013;19(21):5867–5878. doi:10.1158/1078-0432.CCR-12-3753
27. Ji Q, Guo S. Recent advances in TMEM16A: structure, function, and disease. J Cell Physiol. 2019;234(6):7856–7873. doi:10.1002/jcp.27865
28. Li RC, Ke S, Meng FK, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9(8):838. doi:10.1038/s41419-018-0852-y
29. Xu G, Mo L, Wu C, et al. The miR-15a-5p-XIST-CUL3 regulatory axis is important for sepsis-induced acute kidney injury. Ren Fail. 2019;41(1):955–966. doi:10.1080/0886022X.2019.1669460
30. Song J, Zhang P, Liu M, et al. Novel-miR-4885 promotes migration and invasion of esophageal cancer cells through targeting CTNNA2. DNA Cell Biol. 2019;38(2):151–161. doi:10.1089/dna.2018.4377
31. Li W, Wang C, Peng X, Zhang H, Huang H, Liu H. CFTR inhibits the invasion and growth of esophageal cancer cells by inhibiting the expression of NF-κB. Cell Biol Int. 2018;42(12):1680–1687. doi:10.1002/cbin.11069
32. Li Z, Qin X, Bian W, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 2019;38(1):477. doi:10.1186/s13046-019-1473-8
33. Kojima H, Okumura T, Yamaguchi T, Miwa T, Shimada Y, Nagata T. Enhanced cancer stem cell properties of a mitotically quiescent subpopulation of p75NTR-positive cells in esophageal squamous cell carcinoma. Int J Oncol. 2017;51(1):49–62. doi:10.3892/ijo.2017.4001
34. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386. doi:10.1002/ijc.29210
35. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi:10.1016/j.cell.2014.03.008
36. Xu LJ, Yu XJ, Wei B, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci. 2018;22(9):2653–2661. doi:10.26355/eurrev_201805_14961
37. Chen MJ, Deng J, Chen C, Hu W, Yuan YC, Xia ZK. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem Cell Biol. 2019;113:27–36. doi:10.1016/j.biocel.2019.05.011
38. Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25(11):1980–1995. doi:10.1038/s41418-018-0084-9
39. Chen M, Xia Z, Chen C, Hu W, Yuan Y. LncRNA MALAT1 promotes epithelial-to-mesenchymal transition of esophageal cancer through Ezh2-Notch1 signaling pathway. Anticancer Drugs. 2018;29(8):767–773. doi:10.1097/CAD.0000000000000645
40. Su W, Guo C, Wang L, et al. LncRNA MIR22HG abrogation inhibits proliferation and induces apoptosis in esophageal adenocarcinoma cells via activation of the STAT3/c-Myc/FAK signaling. Aging. 2019;11(13):4587–4596. doi:10.18632/aging.102071
41. Hu M, Zhang Q, Tian XH, Wang JL, Niu YX, Li G. lncRNA CCAT1 is a biomarker for the proliferation and drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1 axis. Mol Carcinog. 2019;58(12):2207–2217. doi:10.1002/mc.23109
42. Xu Y, Li Y, Jin J, et al. LncRNA PVT1 up-regulation is a poor prognosticator and serves as a therapeutic target in esophageal adenocarcinoma. Mol Cancer. 2019;18(1):141. doi:10.1186/s12943-019-1064-5
43. Li X, Yang H, Wang J, et al. High level of lncRNA H19 expression is associated with shorter survival in esophageal squamous cell cancer patients. Pathol Res Pract. 2019;215(11):152638. doi:10.1016/j.prp.2019.152638
44. Fan Q, Liu B. Identification of a RNA-seq based 8-long non-coding RNA signature predicting survival in esophageal cancer. Med Sci Monit. 2016;22:5163–5172. doi:10.12659/MSM.902615
45. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
46. Zhang G, Li S, Lu J, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17(1):87. doi:10.1186/s12943-018-0829-6
47. Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin (Shanghai). 2017;49(7):588–597. doi:10.1093/abbs/gmx047
48. Bayraktar R, Pichler M, Kanlikilicer P, et al. MicroRNA 603 acts as a tumor suppressor and inhibits triple-negative breast cancer tumorigenesis by targeting elongation factor 2 kinase. Oncotarget. 2017;8(7):11641–11658. doi:10.18632/oncotarget.14264
49. Wang Y, Zhao YR, Zhang AY, Ma J, Wang ZZ, Zhang X. Targeting of miR-20a against CFLAR to potentiate TRAIL-induced apoptotic sensitivity in HepG2 cells. Eur Rev Med Pharmacol Sci. 2017;21(9):2087–2097.
50. Jiang Y, Cai Y, Shao W, et al. MicroRNA‑144 suppresses aggressive phenotypes of tumor cells by targeting ANO1 in colorectal cancer. Oncol Rep. 2019;41(4):2361–2370. doi:10.3892/or.2019.7025
51. Park YR, Lee ST, Kim SL, et al. Down-regulation of miR-9 promotes epithelial mesenchymal transition via regulating anoctamin-1 (ANO1) in CRC cells. Cancer Genet. 2019;231–232:22–31. doi:10.1016/j.cancergen.2018.12.004
52. Cao Q, Liu F, Ji K, et al. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017;36(1):29. doi:10.1186/s13046-017-0499-z
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.