Back to Journals » Journal of Inflammation Research » Volume 15
LncRNA GAS5 rs145204276 Polymorphism Reduces Renal Cell Carcinoma Susceptibility in Southern Chinese Population
Authors Xiang X, Chen L, He J, Ma G, Li Y
Received 9 November 2021
Accepted for publication 4 February 2022
Published 18 February 2022 Volume 2022:15 Pages 1147—1158
DOI https://doi.org/10.2147/JIR.S348628
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Xiaoyao Xiang,1 Linfa Chen,2 Jiawen He,3 Guoda Ma,3,4 You Li3
1Department of Urology, The Second Affiliated Hospital of Guangdong Medical University, Zhanjiang, 524001, People’s Republic of China; 2Department of NeUrology, Huizhou Third People’s Hospital, Guangzhou Medical University, Huizhou, 516000, People’s Republic of China; 3Guangdong Key Laboratory of Age-Related Cardiac and Cerebral Diseases, Affiliated Hospital of Guangdong Medical University, Zhanjiang, 524001, People’s Republic of China; 4Maternal and Children’s Health Research Institute, Shunde Maternal and Children’s Hospital, Guangdong Medical University, Shunde, 528300, People’s Republic of China
Correspondence: Guoda Ma, Maternal and Children’s Health Research Institute, Shunde Maternal and Children’s Hospital, Guangdong Medical University, Shunde, 528300, People’s Republic of China, Email [email protected] You Li, Guangdong Key Laboratory of Age-Related Cardiac and Cerebral Diseases, Affiliated Hospital of Guangdong Medical University, Zhanjiang, 524001, People’s Republic of China, Email [email protected]
Objective: Recent studies have demonstrated that the long non-coding RNA (lncRNA) GAS5 is closely associated with the onset and progression of several tumor types, including renal cell carcinoma (RCC). This study sought to evaluate the relationship between two functional GAS5 polymorphisms (rs145204276 and rs55829688) and the risk for RCC in the Han Chinese population.
Methods: The rs145204276 and rs55829688 polymorphisms in the GAS5 promoter region were genotyped in 624 RCC patients and 655 age/sex-matched healthy participants. The association between these polymorphisms and RCC risk was then evaluated using odds ratios (ORs) and corresponding 95% confidence intervals (CIs). Additionally, quantitative RT-PCR was used to determine whether these polymorphisms were associated with changes in the levels of expression of GAS5 in 58 RCC patients.
Results: There were significant differences in the GAS5 rs145204276 polymorphism genotype and allele frequencies between the RCC patients and controls (adjusted OR = 0.73, 95% CI = 0.61- 0.87, P = 1.8× 10− 3). When the study participants were stratified based on age, sex, BMI index, and smoking and drinking history, we found that the rs145204276 del allele was associated with a reduced risk for RCC in nondrinkers (P = 3.3× 10− 3), nonsmokers (P = 3.3× 10− 3), females (P = 3.8× 10− 3), and those who were less than 60 years old (P = 3.3× 10− 3). There was also a significant association between the rs145204276 del allele and elevated expression of GAS5 in RCC patients (P = 0.030).
Conclusions: The results of this study revealed an association between the rs145204276 polymorphism in the GAS5 lncRNA and the risk for the development of RCC, thus representing a potentially viable biomarker for identifying individuals at risk of developing this form of cancer.
Keywords: lncRNA GAS5, polymorphism, renal cell carcinoma, case-control
Introduction
Renal cell carcinoma (RCC) accounts for over 90% of all kidney cancers.1 Clear cell RCC is the most common and aggressive form of this disease, with a poor prognosis for the affected patients.2 Although RCC is a relatively common form of cancer, its underlying molecular mechanism is not clear, and both environmental and genetic factors are suspected to be involved in its onset.3 Risk factors associated with RCC include age, sex, alcohol/tobacco use, obesity, and a history of chronic kidney disease.4,5 Additionally, genome-wide association studies (GWAS) have revealed several genetic determinants of RCC in western populations, with single nucleotide polymorphisms (SNPs) in EPAS1, HIF-2α, CCND1, and ITPR2 linked with increased susceptibility to RCC.6–9 However, these SNPs account for less than half of all genetic factors Related to RCC, and the remaining factors need to be identified to increase our understanding of RCC development.
Long non-coding RNAs (lncRNAs) are RNA molecules that have more than 200 nucleotides and that lack protein-coding potential. However, they can regulate gene expression by modulating transcriptional and post-transcriptional factors or by modifying chromatin.10,11 As complex regulatory molecules, lncRNAs are closely associated with key cellular processes, such as inflammation, apoptosis, proliferation, and oncogenesis.12 Also, dysregulation of lncRNA can result in the growth and metastasis of certain tumor types.13–16 Several studies have shown that the growth arrest-specific 5 (GAS5) lncRNA can induce cell cycle arrest and consequent cellular apoptosis in several tissue types.17,18 Thus, recent studies have highlighted the role of GAS5 as a tumor suppressor in osteosarcoma as well as in breast, bladder, and colorectal cancers.19–23 The downregulation of GAS5 has also been linked to the development and progression of RCC.24–26 However, the molecular basis for the downregulation of GAS5 in RCC needs to be explored.
GAS5 is located on chromosome 1q25.1, within a gene containing 12 exons across a 4.087 kb region, coding for 29 different GAS5 splicing variants.27 Mutations within the GAS5 promoter region are known to be linked to oncogenesis. Li et al revealed that the rs145204276 indel polymorphism in the GAS5 promoter region altered the expression of GAS5, and thus protected against the development of gastric cancer.28 Tao et al found that the rs145204276 polymorphism altered the transcriptional activity of GAS5, thereby resulting in an increased risk for hepatocellular carcinoma.29 Wang et al showed that the GAS5 rs55829688 promoter polymorphism altered the binding ability of the YY1 transcription factor to the promoter region, thereby resulting in increased expression of GAS5 and an elevated risk for colorectal cancer.30 Also, Rakhshan et al found that in the Iranian population, the GAS5 rs2067079 polymorphism was linked to an elevated risk for bladder cancer.31 Currently, however, there are no reports on the link between GAS5 promoter region polymorphisms and risk for RCC. Here, we utilized a case-control approach to examine the relationship between such variants and RCC incidence in the Chinese population.
Materials and Methods
Study Participants
This study was performed in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the Second Affiliated Hospital of Guangdong Medical University. All participants provided written informed consent before taking part in the study. We randomly recruited 624 patients with RCC and 655 healthy age/sex-matched control participants from the Second Affiliated Hospital of Guangdong Medical University from September 2014 to August 2020. Patients with RCC had been independently diagnosed by two independent pathologists based on histopathological findings, and partial nephrectomy was performed. None of the patients enrolled in this study had undergone radiotherapy or chemotherapy before participation, and none had any history of previous malignancy. For smoking status, those patients who had smoked less than one cigarette per day for less than 1 year of their life were classified as nonsmokers, and those who had drunk alcohol at least three times per week for longer than 6 months were classified as drinkers. The tumor stage was determined using the international tumor-node-metastasis (TNM) classification system32 and graded according to the 2016 WHO Classification of Tumors of the Urinary System of the Tumor, Nodes and Metastasis system.33 Furthermore, tumors were separated into localized (stage I/II) and advanced (stage III/IV) groups as per the Fuhrman scale based on the criteria of the American Joint Committee on Cancer (AJCC).32,34
Genotyping
We extracted gDNA from the peripheral blood lymphocytes using a genomic DNA purification kit (Qiagen, Guangzhou, China). Two functional polymorphisms of GAS5 lncRNA were selected based on previous reports.29,35 Polymerase chain reaction (PCR) was used for the genotyping of rs145204276 and rs55829688 as described previously.36 The primers for the amplification of rs145204276 were (F: 5’- TCCCGACTGAGGAGGAAGAGCA-3’; R: 5’-AACACCGTCCCGGAAGTGAAA-3’). The PCR products were separated using 7% native polyacrylamide gel electrophoresis and were visualized by silver staining. The genotypes were designated as ins/ins (II), del/ins (ID), or del/del (DD) for each individual. Primers used for the amplification of rs55829688 were: (F: 5’-TGGCTTAGAAGTCCCAGTCA-3’; R: 5’-CGTCCCGGAAGTGAAATCC-3’). Next, the restriction fragment length polymorphism (RFLP) analysis was performed on the amplified PCR products using restriction enzyme BsrDI (NEB, Beijing, China). For quality control, duplicate sequencing of 10% of samples was performed, with a 100% concordance rate.
Quantitative Real-Time PCR (qRT-PCR)
The resected tumor tissues and the adjacent control tissues were obtained from 58 patients with RCC during surgery. None of the patients had undergone radiotherapy or chemotherapy before this operation. The qRT-PCR was performed as we have described previously.37 Total RNA from the control and RCC tumor tissues was extracted using a commercial kit following the manufacturer’s instructions (TianGen, Beijing, China). The RNA concentration, purity, and integrity were determined using a Nano‐Drop ND‐1000 spectrophotometer and 1% agarose gel electrophoresis. Total RNA (1 µg) was converted to cDNA using the RevertAid cDNA Synthesis Kit (Thermo Fisher, Beijing, China). Then, the cDNA was diluted to a final concentration of 10 ng/µL. The resulting cDNAs (1µL; 10 ng) were used as templates to determine the quantity of GAS5 lncRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using quantitative real-time PCR (qRT-PCR) following the SYBR green method. qRT-PCR was performed on a Roche Light Cycler 480 machine with the following pair of primers: 5′- CTTC TGGGCTCAAGTGATCCT-3′ and 5′- TTGTGCCATGAGACTCCATCAG-3′. GAPDH was used as a control for normalization, using the following pair of primers: 5′-GTCAACGGATTTGGTCTGTATT-3′ and 5′-AGTCTTCTGGGTGGCAGTGAT-3′. The qRT-PCR settings were as follows: 95°C for 10 min; 40 cycles at 95°C for 10s, 60°C for 20s, and 72°C for 10s. The 2−ΔΔCt method38 was used to assess the relative expression of GAS5, and all reactions were performed in triplicate.
Cell Culture and Dual-Luciferase Reporter Assay
Human RCC cell line A498 (derived from papillary RCC) and a nonmalignant renal cell line HK-2 were obtained from the American Type Culture Collection (Manassas, VA, USA). The cell lines were incubated in DMEM supplemented with 10% fetal bovine serum and maintained in a humidified incubator (5% CO2) at 37 °C. The lncRNA GAS5 promoter fragment about 300 bp containing the rs145204276 ins allele was synthesized and cloned into the pGL3-basic vector (Promega, WI, USA), yielding the pGL3-ins constructs. The mutant type constructs harbouring the the rs145204276 del allele were generated using QuikChange Lightening Site-Directed Mutagenesis Kit (Stratagene, CA, USA). The resulting constructs were confirmed by direct sequencing. These constructs were transfected into A498 and HK-2 cells using Lipofectamine 3000 (Thermo Fisher, MA, USA). At 24 hours post-transfection, transcriptional activities were determined using the dual-luciferase reporter assay system (Promega, WI, USA) and measurements were presented as the ratio of the firefly and renilla luciferase activities.
Statistical Analysis
SPSS v19.0 (IBM Corp., Armonk, NY, USA) software was used for all statistical analyses. Chi-squared tests were used to test for Hardy-Weinberg equilibrium (HWE), and to analyze the categorical data. Continuous data were analyzed via the Student’s t-tests. The relationship between specific polymorphisms and risk for RCC was assessed using multivariate logistic regression to calculate odds ratios (ORs) and 95% confidence intervals (CIs) after adjusting for age, sex, BMI, smoking status, and drinking status. Mann–Whitney U-tests were used to compare the levels of expression of GAS5 between groups of different genotypes. Relative luciferase activity among groups was compared by Student’s t-tests. P<0.05 was considered statistically significant.
Results
Demographic Data
Table 1 presents the demographic and clinical data for the study participants. There was a similar distribution in terms of age, sex, BMI, smoking status, and drinking status between the RCC and the control groups (P > 0.05).
 |
Table 1 Characteristics of Subjects Between RCC Patients and Control Group |
The Relationship Between GAS5 Polymorphisms and RCC Susceptibility
Table 2 presents the GAS5 polymorphism allele and genotype frequencies for RCC and the control group. No selection bias was evident in either group based on the results of the Hardy-Weinberg equilibrium test (P > 0.05). However, the distribution of rs145204276 polymorphism was significantly different between these two groups (P = 4.8×10−3). There was no significant difference between the RCC and the control group when using a recessive model (II + ID vs DD; P = 0.13). However, in a dominant model (II vs ID + DD), the RCC and the control group differed significantly (P = 1.8×10−3, OR = 0.67, 95% CI: 0.54–0.84). The participants carrying the rs145204276 del allele also exhibited a reduced risk for RCC relative to carriers of the ins allele (P = 1.8×10−3, OR = 0.73, 95% CI: 0.61–0.87). On the contrary, there was a similar distribution of the allele and genotypes for the rs55829688 polymorphism between the RCC patient and the control groups (P > 0.05).
 |
Table 2 Distributions of lncRNA GAS5 Genotypic and Allelic Frequencies Among RCC Patients and Controls |
Subgroup Analyses of GAS5 Polymorphism Distributions According to Participant Clinicopathological Characteristics
Next, we examined the relationships between the GAS5 polymorphisms and the defined clinicopathological variables in the RCC and control groups (Tables 3 and 4). After the stratification of participants based on their age, sex, BMI, drinking status, and smoking status, we found that the rs145204276 del allele was linked with a lower risk for RCC in individuals less than 60 years old (P = 3.3×10−3), female (P = 3.8×10−3), nonsmokers (P = 3.3×10−3) and nondrinkers (P = 3.3×10−3) (Table 3). None of the subgroup analyses revealed any significant differences in the distribution of rs55829688 polymorphism (Table 4).
 |
Table 3 Stratified Analysis Between the Genotypes and Alleles of lncRNA GAS5 rs145204276 Polymorphism Among RCC Patients and Control Group |
 |
Table 4 Stratified Analysis Between the Genotypes and Alleles of lncRNA GAS5 rs55829688 Polymorphism Among RCC Patients and Control Group |
The Relationship Between GAS5 Polymorphisms and RCC Disease Progression
When the relationship between the GAS5 polymorphisms and the clinicopathological characteristics of RCC patients were examined, we found no significant differences in the distribution of polymorphisms, when comparing patients with stage I, II, or III disease, or when comparing patients with moderately (grade I and II) or poorly differentiated (grade III and IV) nuclear grades (Table 5). However, there were significantly fewer individuals having the rs145204276 ID+DD genotype among patients with stage IV disease compared with individuals with the II genotype (P = 0.014, adjusted OR = 0.50, 95% CI: 0.29–0.88). In contrast, no statistical correlations were found among the rs55829688 variant and the RCC stages (P > 0.05).
 |
Table 5 Association Between LncRNA GAS5 Genotypes and Clinicopathologic Characteristics of RCC |
Tissue Level Expression of GAS5
Finally, we examined the relationship between GAS5 polymorphisms, risks for RCC, and GAS5 expression (Figure 1). We found that patient tumor tissue samples exhibited significantly reduced GAS5 expression compared with control tissue samples (P < 0.01) (Figure 1A). Additionally, when stratifying tumor samples based on rs145204276 genotypes, we observed significantly elevated expression of GAS5 in individuals with the rs145204276 ID+DD genotypes compared with individuals with the II genotype (P = 0.030) (Figure 1B). When comparing the rs145204276 ID+DD and II genotypes, no significant difference in the expression of GAS5 was observed in the control samples (P > 0.05) (Figure 1B). In contrast, no statistical difference in GAS5 expression was found among either case or control samples when comparing the rs55829688 TC+CC and TT genotypes (P > 0.05) (Figure 1C).
The Effect of rs145204276 on GAS5 Transcription Activity
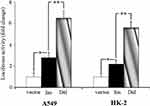
To determine whether rs145204276 alters transcriptional activity, we performed a dual luciferase reporter assay to examine the effects of rs145204276 AGGCA/- polymorphism on the promoter activity. As shown in Figure 2, the luciferase activity of the cells transfected with pGL3-ins or pGL3-del was significantly higher than those of cells transfected with control vectors. In addition, a significant increase of luciferase activity was observed in cells transfected with pGL3-del compared to cells transfected with pGL3-ins in either A498 or HK-2 cell lines (P < 0.01).
Discussion
This study provides novel evidence for the presence of a significant relationship between the GAS5 rs145204276 indel polymorphism and the risk for RCC in a subset of the Chinese population. Additional subgroup analyses further revealed that those who are less than 60 years old, female, nonsmokers or nondrinkers and who carried the rs145204276 del allele might have a lower risk of developing RCC. Additionally, we detected significantly elevated levels of expression of GAS5 in the tumor tissue samples from RCC patients carrying the rs145204276 del allele. However, no significant differences in rs55829688 genotype or allele frequencies were detected when comparing the RCC patients and healthy participants. Thus, there exists a possibility that the GAS5 rs145204276 polymorphism may be a valuable biomarker for predicting the risk for RCC in certain populations.
In the past, several studies have highlighted the relationship between dysregulation of GAS5 and the onset of numerous cancer types, including lung,39 colorectal,40 breast,41 gastric,42 prostate,43 pancreatic,44 and bladder22 cancers. However, the relationship between dysregulation of GAS5 and RCC incidence has not been well studied. Qiao et al observed a significant downregulation of GAS5 in RCC tumor tissue samples and found that this reduction was directly associated with the development and progression of RCC.24 They further found that the overexpression of GAS5 drove tumor cell apoptosis and cell cycle arrest while simultaneously impairing metastasis, thus suggesting that targeting GAS5 may be a viable therapeutic strategy for RCC.24 Toraih et al suggested that the co-expression of GAS5 and miR-34 might act as a biomarker for RCC.27 Thus, these reports indicated a mechanistic role of GAS5 in the development of RCC. However, there were no reports on the involvement of specific alleles of GAS5 that were associated with RCC incidence or progression. Previously, Tao et al had identified the 5-bp indel rs145204276 polymorphism in the GAS5 promoter region, which they found was associated with an elevated risk for hepatocellular carcinoma (HCC).29 In contrast, Zheng et al found that the same rs145204276 polymorphism was linked with reduced risk for colorectal cancer (CRC) and reduced rates of lymph node metastasis in patients with CRC.20 These contradictory findings suggest that this rs145204276 polymorphism may play a tumor type-specific role in the onset and progression of the disease. In this study, we found that there was also a relationship between the GAS5 rs145204276 indel polymorphism and a reduced risk for RCC. Previously, Wang et al had reported that GAS5 promoter region rs55829688 polymorphism was linked with a reduced risk for CRC;45 however, in the present study, we found no significant relationship between the allele frequencies of rs55829688 polymorphism and risk for RCC.
In most patients, the development of RCC is thought to result from a confluence of complex interactions between the genetic and environmental variables; however, these interactions are not completely understood. There is epidemiological evidence, which indicates that there is a strong association between smoking, obesity, hypertension, chronic kidney disease and an elevated risk for RCC.46 Smoking, in particular, is independently associated with a risk for RCC, with a direct relationship between smoking duration and risk for RCC that only declines following smoking cessation.47 The smoking frequency has also been linked to both the onset as well as the development of more advanced RCC.48 Smoking results in immunosuppression, which allows the nascent RCC cells to escape immune detection, thus resulting in an increase in RCC frequency among smokers.49 Epidemiological studies also show a significant relationship between age and RCC incidence, with older individuals exhibiting significantly increased rates of RCC.46 Consistent with these results, the subgroup analyses of our participant cohorts revealed that younger individuals and nonsmokers of particular genotypes were at a significantly lower risk for RCC. Thus, the risk for developing RCC was derived from complex interactions between the environmental and genetic factors.
Functional lncRNA SNPs induce changes in either the expression or the function of these lncRNAs, thereby influencing their interactions with target genes, and potentially influencing disease risk. The GAS5 promoter region rs145204276 indel polymorphism is known to modulate GAS5 expression due to alterations in methylation in the GAS5 promoter region.29,50 Tang et al found that the rs145204276 del allele was bound by the SP1 transcription factor, leading to enhanced promoter activity and consequently elevated expression of GAS5 that resulted in an elevated risk for breast cancer.51 Similarly, Yuan et al found that GAS5 indel polymorphisms were associated with impaired binding of TFAP2A to the promoter region, leading to increased expression of GAS5, which is, in turn, was associated with an elevated risk for gliomas.52 Several other reports showed that the GAS5 rs55829688 T>C polymorphism altered YY1 binding to the promoter region, thereby impairing the expression of GAS5 and reducing the risk for the development of colorectal cancer.45 Yan et al found the rs55829688 polymorphism was linked to elevated TP63 binding to this region, leading to elevated expression of GAS5 that was associated with the development of acute myeloid leukemia (AML).53 In this study, we found that patients with the rs145204276 del allele exhibited an elevated expression of GAS5 in the RCC tissues. It is highly likely that the rs145204276 del allele was associated with higher transcriptional activity, thereby leading to altered expression of GAS5 and a corresponding increased risk for RCC.
This study had several limitations. This was a retrospective analysis conducted in a Hospital; thus, there was an inherent risk of selection bias. Additionally, this study had a relatively limited sample size, restricting its statistical power. Furthermore, due to the lack of access to detailed follow-up data from the study participants, we were not able to fully explore the relationship between these polymorphisms and the prognosis of the RCC patients. Finally, we need to further explore the specific mechanisms involving the modification of the expression of GAS5 due to these GAS5 polymorphisms. It is also necessary to independently validate these results and determine whether they are relevant in populations with other ethnic backgrounds.
Thus, the results of this study provide novel evidence that the GAS5 promoter region rs145204276 indel polymorphism is linked with a reduced risk for RCC in the Han Chinese population. We found that the rs145204276 del allele was associated with elevated levels of expression of GAS5 in this patient population, thereby potentially protecting against the development of RCC. However, further Research is necessary to validate these results and to fully explore how this functional rs145204276 polymorphism would influence the onset and progression of RCC.
Acknowledgments
This work was supported by funding from the National Nature Science Foundation of China (grant numbers 81571157, 81300929 and 81670252) and the Natural Science Foundation of Guangdong Province (grant numbers 2019A1515011424 and 2019A1515011306).
Author Contributions
All authors have made significant contributions to the work reported, whether in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors have no actual or potential conflicts of interest related to this manuscript.
References
1. Kapoor A. Kidney cancer, ESMO 2016. Can Urol Assoc J. 2016;10(11–12Suppl6):S227–S230. doi:10.5489/cuaj.4282
2. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi:10.1038/nrdp.2017.9
3. Semenza JC, Ziogas A, Largent J, Peel D, Anton-Culver H. Gene-environment interactions in renal cell carcinoma. Am J Epidemiol. 2001;153(9):851–859. doi:10.1093/aje/153.9.851
4. Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7(5):245–257. doi:10.1038/nrurol.2010.46
5. Nicodemus KK, Sweeney C, Folsom AR. Evaluation of dietary, Medical and lifestyle risk factors for incident kidney cancer in postmenopausal women. Int J Cancer. 2004;108(1):115–121. doi:10.1002/ijc.11532
6. Gudmundsson J, Sulem P, Gudbjartsson DF, et al. A common variant at 8q24.21 is associated with renal cell cancer. Nat Commun. 2013;4:2776. doi:10.1038/ncomms3776
7. Purdue MP, Johansson M, Zelenika D, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43(1):60–65. doi:10.1038/ng.723
8. Schodel J, Bardella C, Sciesielski LK, et al. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet. 2012;44(4):
9. Wu X, Scelo G, Purdue MP, et al. A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet. 2012;21(2):456–462. doi:10.1093/hmg/ddr479
10. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi:10.1038/nsmb.2480
11. Chen Y, Zhou J. LncRNAs: macromolecules with big roles in neurobiology and neurological Diseases. Metab Brain Dis. 2017;32(2):281–291. doi:10.1007/s11011-017-9965-8
12. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi:10.1016/j.canlet.2013.06.013
13. Zhang Y, Yang R, Lian J, Xu H. LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res. 2016;8(11):5035–5043.
14. Xu E, Yu X, Zeng Q, et al. Functional role of lncRNA DB327252 in lung cancer. J Thorac Dis. 2016;8(10):2793–2802. doi:10.21037/jtd.2016.10.44
15. Xia H, Chen Q, Chen Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7(35):56209–56218. doi:10.18632/oncotarget.10941
16. Taheri M, Omrani MD, Ghafouri-Fard S. Long non-coding RNAs expression in renal cell carcinoma. J Biol Today’s World. 2017;6(12):240–247.
17. Zhang H, Guo Y, Song Y, Shang C. Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother Pharmacol. 2017;79(1):49–55. doi:10.1007/s00280-016-3194-4
18. Mahdi Eftekharian M, Noroozi R, Komaki A, Mazdeh M, Taheri M, Ghafouri-Fard S. GAS5 genomic variants and risk of multiple sclerosis. Neurosci Lett. 2019;701:54–57. doi:10.1016/j.neulet.2019.02.028
19. Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem. 2017;118(12):4772–4781. doi:10.1002/jcb.26145
20. Zheng Y, Song D, Xiao K, et al. LncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7(50):83727–83734. doi:10.18632/oncotarget.13384
21. Li W, Zhai L, Wang H, et al. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7(19):27778–27786. doi:10.18632/oncotarget.8413
22. Liu Z, Wang W, Jiang J, et al. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS One. 2013;8(9):e73991. doi:10.1371/journal.pone.0073991
23. Ghaforui-Fard S, Taheri M. Growth arrest specific transcript 5 in tumorigenesis process: an update on the expression pattern and genomic variants. Biomed Pharmacother. 2019;112:108723. doi:10.1016/j.biopha.2019.108723
24. Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14(2):1077–1082. doi:10.7314/APJCP.2013.14.2.1077
25. Liu L, Pang X, Shang W, Xie H, Feng Y, Feng G. Long non-coding RNA GAS5 sensitizes renal cell carcinoma to sorafenib via miR-21/SOX5 pathway. Cell Cycle. 2019;18(3):257–263. doi:10.1080/15384101.2018.1475826
26. Zhou S, Wang J, Zhang Z. An emerging understanding of long noncoding RNAs in kidney cancer. J Cancer Res Clin Oncol. 2014;140(12):1989–1995. doi:10.1007/s00432-014-1699-y
27. Toraih EA, Alghamdi SA, El-Wazir A, et al. Dual biomarkers long non-coding RNA GAS5 and microRNA-34a co-expression signature in common solid tumors. PLoS One. 2018;13(10):e0198231. doi:10.1371/journal.pone.0198231
28. Li Q, Ma G, Sun S, Xu Y, Wang B. Polymorphism in the promoter region of lncRNA GAS5 is functionally associated with the risk of gastric cancer. Clin Res Hepatol Gastroenterol. 2018;42(5):478–482. doi:10.1016/j.clinre.2018.01.006
29. Tao R, Hu S, Wang S, et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis. 2015;36(10):1136–1143. doi:10.1093/carcin/bgv099
30. Yu J, Ding C, Guan S, Wang C. Association of single nucleotide polymorphisms in the RAB5B gene 3ʹUTR region with polycystic ovary syndrome in Chinese Han women. Biosci Rep. 2019;39:5. doi:10.1042/BSR20190292
31. Rakhshan A, Esmaeili MH, Kahaei MS, et al. A single nucleotide polymorphism in GAS5 lncRNA is associated with risk of bladder cancer in Iranian Population. Pathol Oncol Res. 2019;26(2):1251–1254. doi:10.1007/s12253-019-00693-2
32. Kim SP, Alt AL, Weight CJ, et al. Independent validation of the 2010 American Joint Committee on Cancer TNM classification for renal cell carcinoma: results from a large, single institution cohort. J Urol. 2011;185(6):2035–2039. doi:10.1016/j.juro.2011.02.059
33. Bezhanova SD. [Tumors of the kidney. The new 2016 WHO classification of tumors of the genitourinary system]. Arkh Patol. 2017;79(2):48–52. Russian. doi:10.17116/patol201779248-52
34. Delahunt B, Egevad L, Samaratunga H, Martignoni G, Nacey JN, Srigley JR. Gleason and Fuhrman no longer make the grade. Histopathology. 2016;68(4):475–481. doi:10.1111/his.12803
35. Zhu L, Zhu Q, Wen H, Huang X, Zheng G. Mutations in GAS5 affect the transformation from benign prostate proliferation to aggressive prostate cancer by affecting the transcription efficiency of GAS5. J Cell Physiol. 2019;234(6):8928–8940. doi:10.1002/jcp.27561
36. Moradi M, Gharesouran J, Ghafouri-Fard S, et al. Role of NR3C1 and GAS5 genes polymorphisms in multiple sclerosis. Int J Neurosci. 2020;130(4):407–412. doi:10.1080/00207454.2019.1694019
37. Chen L, Hu W, Li S, et al. Genetic variants of ADAMTS7 confer risk for ischaemic stroke in the Chinese population. Aging (Albany NY). 2019;11(16):6569–6583. doi:10.18632/aging.102211
38. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
39. Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E1–E12. doi:10.1002/mc.22120
40. Liu L, Meng T, Yang XH, et al. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22(2):283–299. doi:10.3233/CBM-171011
41. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi:10.1038/onc.2008.373
42. Sun M, Jin FY, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi:10.1186/1471-2407-14-319
43. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832(10):1613–1623. doi:10.1016/j.bbadis.2013.05.005
44. Lu X, Fang Y, Wang Z, et al. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013;354(3):891–896. doi:10.1007/s00441-013-1711-x
45. Wang Y, Wu S, Yang X, Li X, Chen R. Association between polymorphism in the promoter region of lncRNA GAS5 and the risk of colorectal cancer. Biosci Rep. 2019;39:4.
46. Capitanio U, Bensalah K, Bex A, et al. Epidemiology of Renal Cell Carcinoma. Eur Urol. 2019;75(1):74–84. doi:10.1016/j.eururo.2018.08.036
47. Macleod LC, Hotaling JM, Wright JL, et al. Risk factors for renal cell carcinoma in the VITAL study. J Urol. 2013;190(5):1657–1661. doi:10.1016/j.juro.2013.04.130
48. Lotan Y, Karam JA, Shariat SF, et al. Renal-cell carcinoma risk estimates based on participants in the prostate, lung, colorectal, and ovarian cancer screening trial and national lung screening trial. Urol Oncol. 2016;34(4):167 e169–116. doi:10.1016/j.urolonc.2015.10.011
49. Germano G, Allavena P, Mantovani A. Cytokines as a key component of cancer-related inflammation. Cytokine. 2008;43(3):374–379. doi:10.1016/j.cyto.2008.07.014
50. Xu L, Xia C, Xue B, Sheng F, Xiong J, Wang S. A promoter variant of lncRNA GAS5 is functionally associated with the development of osteosarcoma. J Bone Oncol. 2018;12:23–26. doi:10.1016/j.jbo.2018.03.001
51. Tang Y, Wang Y, Wang X, Liu Y, Zheng K, Genetic A. Variant of rs145204276 in the promoter region of long noncoding RNA GAS5 is associated with a reduced risk of breast cancer. Clin Breast Cancer. 2019;19(3):e415–e421. doi:10.1016/j.clbc.2018.11.006
52. Yuan J, Zhang N, Zheng Y, Chen YD, Liu J, Yang M. LncRNA GAS5 indel genetic polymorphism contributes to glioma risk through interfering binding of transcriptional factor TFAP2A. DNA Cell Biol. 2018;37(9):750–757. doi:10.1089/dna.2018.4215
53. Yan H, Zhang DY, Li X, et al. Long non-coding RNA GAS5 polymorphism predicts a poor prognosis of acute myeloid leukemia in Chinese patients via affecting hematopoietic reconstitution. Leuk Lymphoma. 2017;58(8):1948–1957. doi:10.1080/10428194.2016.1266626
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.