Back to Journals » OncoTargets and Therapy » Volume 9
LncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo
Authors Chen L , Yang H, Xiao Y, Tang X, Li Y, Han Q, Fu J, Yang Y, Zhu Y
Received 13 October 2015
Accepted for publication 5 May 2016
Published 4 July 2016 Volume 2016:9 Pages 4075—4087
DOI https://doi.org/10.2147/OTT.S98203
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Video abstract presented by Long Chen.
Views: 380
Long Chen,1,2,* Huixin Yang,1,* Yanbin Xiao,3,* Xiaoxia Tang,4 Yuqian Li,1 Qiaoqiao Han,1 Junping Fu,3 Yuye Yang,1 Yuechun Zhu1
1Department of Biochemistry and Molecular Biology, Kunming Medical University, Kunming, Yunnan, People’s Republic of China; 2PET/CT Center, Yunan Tumor Hospital, the Third Affiliated Hospital of Kunming Medical University, Kunming Medical University, Kunming, Yunnan, People’s Republic of China; 3Department of Orthopaedics, Yunan Tumor Hospital, the Third Affiliated Hospital of Kunming Medical University, Kunming Medical University, Kunming, Yunnan, People’s Republic of China; 4Department of Pharmacy, the Second Affiliated Hospital of Kunming Medical University, Kunming Medical University, Kunming, Yunnan, People’s Republic of China
*These authors contributed equally to this work
Abstract: The present study intended to demonstrate the effects of long noncoding RNA growth arrest-specific transcript 5 (GAS5) on the migration and invasion of melanoma cells. We first detected the expression of GAS5 among four kinds of melanoma cell lines, followed by constructing GAS5-knocked down and overexpressed stable cells. Next, we evaluated the effects of GAS5 on cell migration and invasion using wound healing and gelatin zymography assays. Finally, melanoma cells with different GAS5 expression were injected into nude mice, and the tumor volumes were recorded and tumor tissues were analyzed after sacrificing the mice. This study systematically examined the function of GAS5 in mediating melanoma metastasis and revealed that GAS5 plays an anticancer role in melanoma via regulating gelatinase A and B, both in vitro and in vivo.
Keywords: lncRNA, GAS5, melanoma, metastasis, gelatinase
Introduction
Long noncoding RNAs (lncRNAs) are RNA molecules that are longer than 200 nucleotides and are not translated into proteins.1 Several investigations have revealed that such molecules play crucial roles in a spectrum of biological processes, especially in neoplasm.2–6 The lncRNA growth arrest-specific 5 (GAS5) was identified and isolated in NIH 3T3 cells in an investigation that aimed to detect genes especially expressed at growth arrest in serum starvation condition.7 The human GAS5-encoding gene comprises 12 exons, with little protein-coding potential, and encodes ten box C/D small nucleolar RNAs (snoRNAs).8 Several studies have shown that GAS5 is downregulated in different kinds of cancers and is correlated with various phenotypes (Table 1). Yet, the expression profile and biological roles of GAS5, especially in migration and metastasis, and its underlying mechanisms in melanoma remain largely unknown.
  | Table 1 GAS5 expression in human cancer |
The incidence of melanoma has been rising worldwide.28–30 Metastasis, which is viewed as a particularly ominous sign, leads to most of the morbidity and mortality in melanoma.31 Moreover, only some of the patients benefit from the new targeted therapies or immunotherapies, and most patients will have to succumb to the metastatic disease in the end. Therefore, understanding the precise mechanism of melanoma as much as possible and developing innovative medicine targeting metastasis remain emergent and meaningful.
Matrix metalloproteinases (MMPs), which can cleave peptide bonds thereby degrading proteins, play crucial roles in a spectrum of diseases, such as rheumatoid arthritis, cardiovascular disease, and chronic obstructive pulmonary disease.32–34 Moreover, MMPs have also been correlated with various tumor metastases,35–37 including melanoma.38–42 Based on the structure and substrate specificity, >20 MMPs are classified into six groups, which are the collagenases, gelatinases, membrane type, stromelysins, matrilysins, and others.
Herein, we focus on the effects of GAS5 on melanoma metastasis and its possible correlation with gelatinase A and B (MMP2 and MMP9).
Materials and methods
Cell culture
Hacat, A375, M21, SK-Mel-28, and SK-Mel-110 cell lines (American Type Culture Collection, Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Hyclone, Logan, UT, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific) at 37°C in 5% (v/v) CO2. Upon reaching 70%–80% confluence, cells were digested by trypsin (Sigma-Aldrich Co., St Louis, MO, USA) and all the cells used in the current study were within three passages. The Ethics Committee of Kunming Medical University did not require the authors to obtain ethics approval for cell/cell line use.
Total RNA extraction and detection of GAS5 by quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cultured cells by using the Trizol reagent (Thermo Fisher Scientific) and treated with RNase-free DNase (Boehringer Ingelheim, Ingelheim, Germany) at 37°C for 40 minutes, followed by phenol–chloroform extraction and ethanol precipitation. Total RNA (2 μg) was used in the reverse transcription reaction with oligo(dT) primers by using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. qRT-PCR was carried out by using FastStart Universal SYBR Green Master (Rox; Hoffman-La Roche Ltd., Basel, Switzerland) with the Applied Biosystems 7300 Real-Time PCR system (Thermo Fisher Scientific). The specificity of the PCR product was confirmed by melting curve analysis and gel electrophoresis. GAS5 gene expression level was normalized to that of the housekeeping gene GAPDH and calculated using the 2-ΔΔCt method. PCR was performed as follows: 5 minutes at 94°C; then 30 cycles of 95°C for 20 seconds, 65°C for 20 seconds, and 72°C for 20 seconds; and then a 5-minute extension at 72°C. The primers used were GAPDH (forward, 5′-GGGCATCTTGGGCTACAC-3′; reverse, 5′-GGTCCAGGGTTTCTTACTCC-3′; 209-bp band product), GAS5 (forward, 5′-AGCTGGAAGTTGAAATGG-3′; reverse, 5′-CAAGCCGACTCTCCATACC-3′; 123-bp band product), MMP2 (forward, 5′-CAGGGAATGAGTACTGGGTCTATT-3′; reverse, 5′-ACTCCAGTTAAAGGCAGCATCTAC-3′; 178-bp band product), and MMP9 (forward, 5′-AATCTCTTCTAGAGACTGGGAAGGAG-3′; reverse, 5′-AGCTGATTGACTAAAGTAGCTGGA-3′; 207-bp band product).
Establishment of stable GAS5 overexpression and knockdown in human melanoma cells
To generate A375-GAS5si and SK-Mel-110-GAS5over stably expressing cell lines, lentivirus was made using a three-plasmid packaging system (Takara, Kusatsu, Japan). A shRNA with high GAS5 knockdown efficiency was used (GAS5siRNA: CTTGCCTGGACCAGCTTAA). The lentiviral vector system contains the pSIREN-RetroQ-ZsGreen1 vector, the pGag/pol, and the pVSVG element vector. shRNAs targeting GAS5 and negative control are harbored in the pSIREN-RetroQ-ZsGreen1 vector. DNA sequencing confirmed the accurate insertion of the GAS5shRNA. A total of 15 μg of pGag/pol, 10 μg of pVSVG, and 20 μg of pSIREN-RetroQ-ZsGreen1-GAS5shRNA plasmid were cotransfected into subconfluent 293T cells using Lipofectamine® 2000 Reagent (Thermo Fisher Scientific). The viral supernatants were collected 72 hours later. Following the transfection of the A375 with this recombinant deficient lentivirus, the clones with green fluorescence were selected under fluorescence microscopy, after which the experimental procedures were performed. Meanwhile, the pLVX-TRE3G-ZsGreen1 vector harboring the GAS5 cDNA (NR_002578.2) lentivirus was harvested 72 hours after transfection. Subconfluent A375 cells were infected with harvested lentivirus and were selected in 2 μg/mL of puromycin for 3 days, and then the retained cells were cultured in 1 μg/mL of puromycin to produce a polyclonal cell population. Individual colonies of stable overexpression of GAS5 and control colonies were isolated.
Wound healing assay
A375, A375-GAS5si, SK-Mel-110, and SK-Mel-110-GAS5over cells were seeded into four six-well plates at a density of 1×105 cells per well, and cultured until 90% confluency (after ~24 hours). A 200 μL pipette tip was used to make a straight scratch, and the wells were washed twice with PBS to elute the floating cells. Next, the media were replaced with serum-free Dulbecco’s Modified Eagle’s Medium containing 2 μg/mL of mitomycin C, which was added at the time of scratch infliction to inhibit the cell division. Reference marks were made across the bottoms of the wells with an ultrafine marker. Images were taken at 0, 12, and 24 hours after scratching using an Olympus IX51 microscope, and migration distances of cells were calculated in a blinded manner by using ImageJ software. Scratch width was subtracted from the time 0 scratch widths at the same location to determine cell migration distance. Migration distances were averaged to determine overall migration.
Gelatin zymography
Gelatin zymography was carried out as described previously.43 In general, conditioned media from the different cell lines were diluted 1:1 in nonreducing sample buffer and separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a 8% polyacrylamide gel containing 1.5 mg/mL gelatin (Cosmo Bio, Tokyo, Japan) for 60 minutes at 85 V, followed by another 60 minutes at 130 V. The gel was renatured by incubation with 2.5% Triton X-100 for 30 minutes at room temperature. The gels were washed for 30 minutes in developing buffer (50 mM Tris–HCl pH 8.0, 2.5 mM CaCl2) overnight at 37°C. Finally, gels were stained with 0.25% Coomassie blue R-250 for 45 minutes and then destained appropriately. Proteolytic bands in the zymography were quantified by scanning densitometry using the ImageJ 1.46 program (United States National Institutes of Health).
Western blotting to detect MMP2 and MMP9 expression
Protein concentration of cell lysates was determined by NanoDrop ND-1000 (Thermo Fisher Scientific), and equal amounts of proteins (10 μg) of cell lysates were separated on a 10% SDS-PAGE and electrotransferred onto a nitrocellulose membrane (EMD Millipore, Billerica, MA, USA). The membranes were blocked in Tris buffered saline Tween containing 5% fat-free dry milk and then incubated with primary antibodies overnight at 4°C, followed by incubation with a horseradish peroxidase-conjugated secondary antibody for 1 hour. Primary antibodies rabbit monoclonal to MMP-2 (1:3,000 dilution, ab192082; Abcam, Cambridge, MA, USA), rabbit monoclonal antibody to MMP-9 (1:1,000 dilution, ab194316; Abcam), and rabbit monoclonal antibody to GAPDH (1:5,000 dilution, ab181602; Abcam) were used. Proteins were detected using enhanced chemiluminescence reagents (EMD Millipore).
Tumor mouse model
All surgical procedures and care administered to the mice were in accordance with Kunming Medical University institutional guidelines. Six-week-old male immunodeficient BALB/c-nu mice were purchased from Beijing HFK Bioscience Co., Ltd (Beijing, People’s Republic of China) and maintained in a barred animal facility at Kunming Medical University. Mice were regularly monitored for any distress once the tumor was established and were humanely sacrificed on reaching the end point. Regular health and well-being checks were also performed by Kunming Medical University veterinarian staff. A375, A375-GAS5si, SK-Mel-110, and SK-Mel-110-GAS5over cells were seeded into 25 cm2 cell culture bottles. After 36-hours incubation, 5×106 cells were subcutaneously injected into the right posterior of mice. Tumor volumes were calculated every 3 days according to the formula, tumor volume (mm3) = length × width2 ×0.5. Mice were euthanized on day 26 and tumors were excised. The Kunming Medical University Animal Care and Use Committee approved all the experimental procedures used for mice.
Statistical analysis
All experiments were performed in triplicate on three independent occasions and data are expressed as mean ± standard deviation, and the difference among all groups was compared by one-way analysis of variance with least significant difference for the comparison between two groups. A P-value of <0.05 was considered significant. All the statistical analyses were carried out with SPSS Version 21.0 (IBM Corporation, Armonk, NY USA).
Results
GAS5 expression varies among melanoma cells
GAS5 gene is located in 1q25.1 and harbored as many as 12 exons and numerous snoRNAs. There are two types of mature GAS5 transcripts, GAS5a and GAS5b, which can be distinguished from the seventh exon (Figure 1A). qRT-PCR was employed to detect the GAS5 expression level among Hacat, A375, M21, SK-Mel-28, and SK-Mel-110 cell lines. Results revealed that compared with Hacat, GAS5 expression varies sharply among different melanoma cell lines. Notably, compared with Hacat, A375 expressed the highest GAS5 expression level (2.19±0.13, P=0.0003), whereas the lowest GAS5 expression was found in SK-Mel-110 cell lines (0.065±0.009, P=0.0002; Figure 1B).
Construction of GAS5 stable overexpression and knockdown melanoma cells
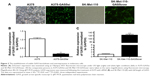
Full length cDNA of GAS5 was synthesized and purified (Genewiz, Suzhou, People’s Republic of China) according to the sequence provided by National Center for Biotechnology Information (NR_002578.2), and the plasmids are supplied in the supplementary materials (Figures S1 and S2). Three shRNAs were designed, and the shRNA with the highest interfering efficiency was chosen to construct recombinant deficient lentivirus to knockdown GAS5 in A375 cells (Figures S3 and S4). The blank and fluorescence images of A375 and SK-Mel-110 cells were taken after being incubated with deficient lentivirus and selected (Figure 2A), followed by detecting GAS5 expression via qRT-PCR. Approximately 90% of GAS5 was knocked down in A375-GAS5si cells, while the GAS5 expression in SK-Mel-110-GAS5over cells is nearly 120 folds more than in SK-Mel-110 cells (P=0.0018 and 0.0007, respectively; Figure 2B and C).
GAS5 inhibits the migration of melanoma cells
To evaluate whether knocking down or overexpressing GAS5 affects the oncogenic behavior of melanoma cells, the migratory ability of melanoma cells were detected. Indeed, the capability to fill the gap of scrape injuries, mechanically made in confluent cell cultures, depends on both cell proliferation and migration, so we treated cells with mitomycin C to inhibit cell proliferation. Closure analysis revealed a significant acceleration in filling the gap for A375-GAS5si, and an obvious decreased ability to fill it in SK-Mel-110-GAS5over cells at 12 hours (P=0.0091 and P<0.0001, respectively; Figure 3).
GAS5 attenuates the activity of gelatinase A and B in melanoma cells
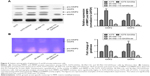
We next detect the MMP2 and MMP9 protein expression level by Western blot and found that knocking down GAS5 upregulated both the MMP2 and MMP9 expression, whereas overexpressing GAS5 significantly decreased both of them (P<0.01; Figure 4A). Moreover, given that the activities of MMPs play crucial roles in tumor metastasis, we next analyzed MMPs in protein lysates by zymography, to clarify whether MMPs are involved in anticancer activity by GAS5. We evaluated the effects of GAS5 on MMP9 and MMP2 activities with the use of gelatin zymography. Quantitative analysis of the detected MMP activities by ImageJ Software confirmed these observations. Based on the different sizes of the proteins, we could detect the proactive and active forms of MMP2 and MMP9 in all analyzed samples. Comparable changes were observed for the total activities of MMP2 and MMP9 as well as the latent proforms of these enzymes that are activated during zymography by SDS-mediated denaturation and subsequent renaturation of the zymogens. The activities of both MMPs showed significant quantitative differences among the control cells. Data revealed that the gelatinolytic activities of MMP2 and MMP9 increased in A375-GAS5si cells, whereas overexpressing GAS5 significantly suppressed these phenomena (P<0.01; Figure 4B).
GAS5 inhibits the tumor growth in vivo
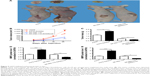
To detect whether GAS5 inhibits tumor growth in vivo, we observed the growth of melanoma tumors in mice formed from the A375, A375-GAS5si, SK-Mel-110, and SK-Mel-110-GAS5over. The eight nude mice developed neoplastic lesions on injection position following the xenograft of these four kinds of melanoma cells. Larger tumors were found in the mice injected with the A375-GAS5si cells compared with those treated with A375 cells (Figure 5A). Meanwhile, smaller tumors were observed in the mice injected with SK-Mel-110-GAS5over cells relative to those treated with SK-Mel-110 cells (Figure 5A). On day 26, all the mice were sacrificed and the tumor mass of each mouse was evaluated. The average tumor size and tumor mass in the mice injected with the A375-GAS5si cells were threefold and 68% higher than those of the tumor in mice injected with A375 cells, respectively (Figure 5B and C). Moreover, the average tumor size and tumor mass in the mice injected with the SK-Mel-110-GAS5over cells reduced by 30% and 40%, respectively, compared with those of the mice treated with SK-Mel-110 (Figure 5B and C). These results indicate that GAS5 functions as a tumor suppressor in melanoma. We next investigated the inhibitory effect of GAS5 on gelatinase expression in tumor tissues harvested from A375, A375-GAS5si, SK-Mel-110, and SK-Mel-110-GAS5over-treated mice. Knocking down GAS5 was observed to significantly increase the MMP2 and MMP9 expression, whereas overexpressing GAS5 downregulated the levels of MMP2 and MMP9 (Figure 5D and E). Collectively, these results clearly demonstrate that GAS5 can be a potent tumor suppressor in this animal model.
Discussion
In the present study, we evaluated the expression of lncRNA GAS5 among melanoma cells and revealed its diverse expression level. By overexpressing GAS5 in SK-Mel-110 cell line in which GAS5 expression is the least, and knocking down GAS5 in A375 cell line where GAS5 expression is mostly abundant, we detected the migratory ability via wound healing assay, and metastatic probability through gelatin zymography, among A375, A375-GAS5si, SK-Mel-110, and SK-Mel-110-GAS5over cells. The results unraveled that knocking down GAS5 facilitated the wound healing and increased the proteolytic potential in A375 cells, whereas overexpressing GAS5 demonstrated the reverse results. We next determined the biological effects of GAS5 on tumor growth in vivo and found that GAS5 significantly inhibits tumor growth. In general, our findings demonstrated that GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo.
Since 1988 when GAS5 was isolated in growth arrest cells by Schneider et al,7 decreased GAS5 levels have been found in a spectrum of solid tumors and blood cancers (Table 1). However, the role of GAS5 in melanoma, a disease with increasing incidence and limited treatment as well as poor prognosis,29,44 is largely unknown. Investigation between GAS5 and melanoma is limited. In the year 2000, Smedley et al45 characterized the chromosome 1 abnormalities in six malignant melanoma cell lines, and they found out that the 1q25.1 region that harbors GAS5 is deleted in WM1433 melanoma cell lines. However, the GAS5 expression among other melanoma cells is still unclear, let alone the biological functions and molecular mechanisms. Based on the fact that GAS5 has been related to a number of cancers (Table 1), and the fact that the origination, development, and metastasis of melanoma have been correlated with considerable noncoding RNAs,46–51 we tentatively hypothesize that there is a further underlying correlation between GAS5 and melanoma. First, we detected the GAS5 expression among Hacat, A375, M21, SK-Mel-28, and SK-Mel-110, and the qRT-PCR results demonstrated that the GAS5 expression varies differently in these melanoma cells, which revealed that GAS5 expression can be influenced not only by species but also by different cell lines of the identical diseases, implying that complicated mechanisms and factors may be involved.
Moreover, given that metastasis is the leading lethal cause of numerous cancers,52–56 including melanoma,31,57,58 we focused our investigation on the effects of GAS5 on melanoma migration and invasion. Knocking down GAS5 significantly speeded the wound healing, whereas overexpressing it retarded this phenomenon, indicating an anticancer tendency of GAS5, which backed up its tumor-inhibiting characteristic.
Many studies have revealed that aberrant MMPs play important roles in tumor initiation, development, migration, invasion, and metastasis, including breast cancer, lung cancer, hepatocarcinoma, and gallbladder carcinoma.59–65 The gelatinase A and B, that is, MMP2 and MMP9, have been unraveled to be closely correlated with the invasive potential and metastasis among a spectrum of cancers including melanoma.66–69 Consistent with these events, here we detected the expression level of MMP2 and MMP9 by Western blotting and found a marked increase in the expression of MMP2 and MMP9 in A375-GAS5si melanoma cells and a significant decrease in SK-Mel-110-GAS5over, demonstrating that GAS5 negatively mediates the expression of both MMP2 and MMP9. Given that there are conflicting reports regarding the expression and activity of MMPs, for instance, Zhang et al70 found that estradiol-I7beta increased MMP-9 activation, whereas decreased MMP-9 expression in the mouse uterus, we carried out the zymography experiments intended to detect the gelatinase activity. The gelatin zymography, first described in 1980 by Heussen and Dowdle, is a commonly used, semiquantitative, and very sensitive technique, which can be used to detect the gelatinase levels.71 We confirmed the phenomenon that the activities of MMP2 and MMP9 were increased in A375-GAS5si cells and the reverse phenomenon in SK-Mel-110-GAS5over. Our results (both Western blotting and gelatin zymography) demonstrated that the anticancer effects of GAS5 are associated with decreased MMPs. Consistent with this study, several previous investigations demonstrated that long noncoding RNAs such as HOTAIR,72 MALAT1,73 and FENDRR74 had anticancer or procancer activity through MMPs. Interestingly, GAS5 has also been correlated with MMPs but in a seemingly converse way. Song et al75 found that inducting the exogenous GAS5 in osteoarthritis chondrocytes resulted in markedly increased activation of MMP2 and MMP9. Although we believe that both the different cell lines and the complexities of cancer possibly contributed to this difference, conversely, Song’s study also identified that GAS5 could interact with MMP2 and MMP9.
We could not collect enough clinical samples to analyze the GAS5 expression, which may better illustrate the GAS5 function and possible mechanisms. In fact, we are collecting melanoma clinical samples and hope to publish our investigations in the future.
Conclusion
Overall, our results demonstrated that GAS5 regulates the metastasis phenotype of melanoma cells via downregulation of proteolytic enzymes MMP2 and MMP9 in vitro and inhibits tumor growth in vivo, providing preclinical support for the therapeutic potential of GAS5 in melanoma management. Therefore, this strategy opens up new opportunities for design of innovative agents with the goal of improving melanoma care and might benefit a subset of melanoma patients by site-directed overexpressing of GAS5.
Acknowledgments
This work was funded by the National Science Foundation of China (number 81160246 and number 81160421) and the Science and Technology Fund of Yunnan Province (number 2013FB102).
Disclosure
The authors report no conflicts of interest in this work.
References
Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13): 1494–1504. | ||
Peng L, Yuan X, Jiang B, Tang Z, Li G-C. LncRNAs: key players and novel insights into cervical cancer. Tumor Biol. 2016;37(3):2779–2788. | ||
Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612. | ||
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. | ||
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. | ||
Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. | ||
Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54(6):787–793. | ||
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. | ||
Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14(2):1077–1082. | ||
Solocinski K, Barilovits S, Welch A, Wingo C, Cain B, Gumz M. Circadian regulation of lncRNAs connects Per1, endothelin-1, and GR. FASEB J. 2015;29(1 suppl):814.10. | ||
Cao Q, Wang N, Qi J, Gu Z, Shen H. Long noncoding RNAGAS5 acts as a tumor suppressor in bladder transitional cell carcinoma via regulation of chemokine (CC motif) ligand 1 expression. Mol Med Rep. 2016;13(1):27–34. | ||
Liu Z, Wang W, Jiang J, et al. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS One. 2013;8(9):e73991. | ||
Tu Z-Q, Li R-J, Mei J-Z, Li X-H. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7(7):4303–4309. | ||
Chang L, Li C, Lan T, et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13(2):1541–1550. | ||
Sun M, Jin F-Y, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14(1):319. | ||
Liu Y, Zhao J, Zhang W, et al. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. | ||
Cao S, Liu W, Li F, Zhao W, Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7(10):6776–6783. | ||
Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(S1):E1–E12. | ||
Dong S, Qu X, Li W, et al. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol. 2015;8(1):43. | ||
Chiang I, Wang W-S, Liu H-C, Yang S-T, Tang N-Y, Chung J-G. Curcumin alters gene expression-associated DNA damage, cell cycle, cell survival and cell migration and invasion in NCI-H460 human lung cancer cells in vitro. Oncol Rep. 2015;34(4):1853–1874. | ||
Lu X, Fang Y, Wang Z, et al. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013;354(3):891–896. | ||
Renganathan A, Kresoja-Rakic J, Echeverry N, et al. GAS5 long non-coding RNA in malignant pleural mesothelioma. Mol Cancer. 2014;13(1):119. | ||
Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):253. | ||
Krell J, Frampton AE, Mirnezami R, et al. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS One. 2014;9(6):e98561. | ||
Williams G, Pickard M. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget. 2016;7(9):10104–10116. | ||
Mourtada-Maarabouni M, Pickard M, Hedge V, Farzaneh F, Williams G. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. | ||
Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat. 2014;145(2):359–370. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Torre LA, Sauer AM, Chen MS Jr, Kagawa-Singer M, Jemal A, Siegel RL. Cancer statistics for Asian Americans, Native Hawaiians, and Pacific Islanders, 2016: converging incidence in males and females. CA Cancer J Clin. 2016;66(3):182–202. | ||
Damsky WE, Theodosakis N, Bosenberg M. Melanoma metastasis: new concepts and evolving paradigms. Oncogene. 2014;33(19):2413–2422. | ||
Goncalves I, Bengtsson E, Colhoun HM, et al. Elevated plasma levels of MMP-12 are associated with atherosclerotic burden and symptomatic cardiovascular disease in subjects with type 2 diabetes. Arterioscler Thromb Vasc Biol. 2015;35(7):1723–1731. | ||
Araki Y, Wada TT, Aizaki Y, et al. Histone methylation and STAT3 differentially regulate IL-6-induced MMP gene activation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheumatol. 2016;68(5):1111–1123. | ||
Papakonstantinou E, Karakiulakis G, Batzios S, et al. Acute exacerbations of COPD are associated with significant activation of matrix metalloproteinase 9 irrespectively of airway obstruction, emphysema and infection. Respir Res. 2015;16:78. | ||
Lee WT, Lee TH, Cheng CH, Chen KC, Chen YC, Lin CW. Antroquinonol from Antrodia camphorata suppresses breast tumor migration/invasion through inhibiting ERK-AP-1- and AKT-NF-kappaB-dependent MMP-9 and epithelial-mesenchymal transition expressions. Food Chem Toxicol. 2015;78:33–41. | ||
Chou YC, Chang MY, Wang MJ, et al. PEITC inhibits human brain glioblastoma GBM 8401 cell migration and invasion through the inhibition of uPA, Rho A, and Ras with inhibition of MMP-2, -7 and -9 gene expression. Oncol Rep. 2015;34(5):2489–2496. | ||
Liu Y, Zheng D, Liu M, et al. Downregulation of glypican-3 expression increases migration, invasion, and tumorigenicity of human ovarian cancer cells. Tumour Biol. 2015;36(10):7997–8006. | ||
Ji BC, Hsiao YP, Tsai CH, et al. Cantharidin impairs cell migration and invasion of A375.S2 human melanoma cells by suppressing MMP-2 and -9 through PI3K/NF-kappaB signaling pathways. Anticancer Res. 2015;35(2):729–738. | ||
Shi H, Liu L, Liu LM, Geng J, Chen L. Inhibition of tumor growth by beta-elemene through downregulation of the expression of uPA, uPAR, MMP-2, and MMP-9 in a murine intraocular melanoma model. Melanoma Res. 2015;25(1):15–21. | ||
Bi MC, Hose N, Xu CL, Zhang C, Sassoon J, Song E. Nonlethal levels of zeaxanthin inhibit cell migration, invasion, and secretion of MMP-2 via NF-kappaB pathway in cultured human uveal melanoma cells. J Ophthalmol. 2016;2016:8734309. | ||
Shoshan E, Braeuer RR, Kamiya T, et al. NFAT1 directly regulates IL-8 and MMP-3 to promote melanoma tumor growth and metastasis. Cancer Res. Epub 2016 Mar 24. | ||
Stasiak M, Boncela J, Perreau C, et al. Lumican inhibits SNAIL-induced melanoma cell migration specifically by blocking MMP-14 activity. PLoS One. 2016;11(3):e0150226. | ||
Hu X, Beeton C. Detection of functional matrix metalloproteinases by zymography. Journal of visualized experiments. 2010;8(45):2445. | ||
Lo JA, Fisher DE. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science. 2014;346(6212):945–949. | ||
Smedley D, Sidhar S, Birdsall S, et al. Characterization of chromosome 1 abnormalities in malignant melanomas. Genes Chromosomes Cancer. 2000;28(1):121–125. | ||
Zhao W, Mazar J, Lee B, et al. The long noncoding RNA SPRIGHTLY regulates cell proliferation in primary human melanocytes. J Invest Dermatol. 2016;136(4):819–828. | ||
Aftab MN, Dinger ME, Perera RJ. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch Biochem Biophys. 2014;563:60–70. | ||
Li R, Zhang L, Jia L, et al. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 2014;9(6):e100893. | ||
Mazar J, Zhao W, Khalil AM, et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5(19):8959–8969. | ||
Tian Y, Zhang X, Hao Y, Fang Z, He Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014;24(4):335–341. | ||
Lessard L, Liu M, Marzese DM, et al. The CASC15 long intergenic noncoding RNA locus is involved in melanoma progression and phenotype switching. J Invest Dermatol. 2015;135(10):2464–2474. | ||
McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16(8):717–727. | ||
Sun Y, Ma L. The emerging molecular machinery and therapeutic targets of metastasis. Trends Pharmacol Sci. 2015;36(6):349–359. | ||
Makker A, Goel MM. Tumor progression, metastasis, and modulators of epithelial-mesenchymal transition in endometrioid endometrial carcinoma: an update. Endocr Relat Cancer. 2016;23(2):R85–R111. | ||
Scott CD, Harpole DH. The biology of pulmonary metastasis. Thorac Surg Clin. 2016;26(1):1–6. | ||
Zhong J, Chen Y, Wang LJ. Emerging molecular basis of hematogenous metastasis in gastric cancer. World J Gastroenterol. 2016;22(8):2434–2440. | ||
AlQathama A, Prieto JM. Natural products with therapeutic potential in melanoma metastasis. Nat Prod Rep. 2015;32(8):1170–1182. | ||
Neagu M, Constantin C, Longo C. Chemokines in the melanoma metastasis biomarkers portrait. J Immunoassay Immunochem. 2015;36(6):559–566. | ||
Stallings-Mann ML, Waldmann J, Zhang Y, et al. Matrix metalloproteinase induction of Rac1b, a key effector of lung cancer progression. Sci Transl Med. 2012;4(142):142ra195. | ||
Porlan E, Marti-Prado B, Morante-Redolat JM, et al. MT5-MMP regulates adult neural stem cell functional quiescence through the cleavage of N-cadherin. Nat Cell Biol. 2014;16(7):629–638. | ||
Chung EY, Ochs CJ, Wang Y, et al. Activatable and cell-penetrable multiplex FRET nanosensor for profiling MT1-MMP activity in single cancer cells. Nano Lett. 2015;15(8):5025–5032. | ||
Garcia-Irigoyen O, Latasa MU, Carotti S, et al. Matrix metalloproteinase 10 contributes to hepatocarcinogenesis in a novel crosstalk with the stromal derived factor 1/C-X-C chemokine receptor 4 axis. Hepatology. 2015;62(1):166–178. | ||
Lee DG, Lee SH, Kim JS, et al. Loss of NDRG2 promotes epithelial-mesenchymal transition of gallbladder carcinoma cells through MMP-19-mediated Slug expression. J Hepatol. 2015;63(6): 1429–1439. | ||
Chevalier C, Collin G, Descamps S, et al. TOM1L1 drives membrane delivery of MT1-MMP to promote ERBB2-induced breast cancer cell invasion. Nat Commun. 2016;7:10765. | ||
Wong HL, Jin G, Cao R, Zhang S, Cao Y, Zhou Z. MT1-MMP sheds LYVE-1 on lymphatic endothelial cells and suppresses VEGF-C production to inhibit lymphangiogenesis. Nat Commun. 2016;7:10824. | ||
Tabouret E, Bertucci F, Pierga JY, et al. MMP2 and MMP9 serum levels are associated with favorable outcome in patients with inflammatory breast cancer treated with bevacizumab-based neoadjuvant chemotherapy in the BEVERLY-2 study. Oncotarget. Epub 2016 Feb 23. | ||
Tabouret E, Boudouresque F, Farina P, et al. MMP2 and MMP9 as candidate biomarkers to monitor bevacizumab therapy in high-grade glioma. Neuro Oncol. 2015;17(8):1174–1176. | ||
Che YL, Luo SJ, Li G, et al. The C3G/Rap1 pathway promotes secretion of MMP-2 and MMP-9 and is involved in serous ovarian cancer metastasis. Cancer Lett. 2015;359(2):241–249. | ||
Poudel B, Lee YM, Kim DK. DDR2 inhibition reduces migration and invasion of murine metastatic melanoma cells by suppressing MMP2/9 expression through ERK/NF-kappaB pathway. Chin J Biochem Biophys. 2015;47(4):292–298. | ||
Zhang X, Christenson LK, Nothnick WB. Regulation of MMP-9 expression and activity in the mouse uterus by estrogen. Mol Reprod Dev. 2007;74(3):321–331. | ||
Heussen C, Dowdle EB. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal Biochem. 1980;102(1):196–202. | ||
Kim HJ, Lee DW, Yim GW, et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46(2):521–530. | ||
Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36(3):1477–1486. | ||
Xu TP, Huang MD, Xia R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. | ||
Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res. 2014;32(12):1628–1635. |
Supplementary materials
  | Figure S2 Sequencing results for the cloned GAS5 sequence in SK-Mel-110 cells. |
  | Figure S4 BLAST analysis of the recombinant GAS5 gene. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.