Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA FTX Contributes to the Progression of Colorectal Cancer Through Regulating miR-192-5p/EIF5A2 Axis
Authors Zhao K, Ye Z, Li Y, Li C, Yang X, Chen Q, Xing C
Received 3 December 2019
Accepted for publication 29 February 2020
Published 31 March 2020 Volume 2020:13 Pages 2677—2688
DOI https://doi.org/10.2147/OTT.S241011
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Kui Zhao,1 Zhenyu Ye,2 Yecheng Li,1 Chunyan Li,3 Xiaodong Yang,1 Qiang Chen,1 Chungen Xing1
1Department of Gastrointestinal Surgery, The Second Affiliated Hospital of Soochow University, Suzhou 215004, Jiangsu, People’s Republic of China; 2Department of Hepatobiliary and Pancreatic Surgery, The Second Affiliated Hospital of Soochow University, Suzhou 215004, Jiangsu, People’s Republic of China; 3Division of Nanobiomedicine, Suzhou Institute of Nano-Tech and Nano-Bionics, Chinese Academy of Sciences, Suzhou 215123, Jiangsu, People’s Republic of China
Correspondence: Chungen Xing
Department of Gastrointestinal Surgery, The Second Affiliated Hospital of Soochow University, No. 1055, Sanxiang Road, Suzhou, Jiangsu 215004, People’s Republic of China
Tel +86 512 67784109
Email [email protected]
Background: Long non-coding RAN five prime to Xist (LncRNA FTX) has been revealed to be a cancer-related lncRNA and implicated in the progression of colorectal cancer (CRC). Besides, miR-192-5p (miR-192) or eukaryotic initiation factor 5A2 (EIF5A2) also was identified to link with the tumorigenesis of CRC. Here, we further explored the function of FTX and the regulatory relationship among FTX, miR-192 and EIF5A2 in CRC progression.
Methods: Levels of FTX, miR-192-5p and EIF5A2 were detected by quantitative real-time polymerase chain reaction (qRT-PCR) or Western blot, respectively. Cell proliferation, apoptosis, migration and invasion were analyzed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, flow cytometry or transwell assay, respectively. The interaction between miR-192-5p and FTX or EIF5A2 was confirmed by dual-luciferase reporter and pull-down assay. Murine xenograft model was established using LoVo cells transfected with sh-FTX.
Results: FTX was up-regulated in CRC tissues and cell lines, knockdown of FTX inhibited CRC cell proliferation, migration and invasion in vitro as well as suppressed CRC tumor growth in vivo. FTX was confirmed to directly bind to miR-192-5p and negatively regulated miR-192-5p expression in CRC cells. Besides that overexpressed FTX positively modulated EIF5A2, a direct target of miR-192-5p, via miR-192-5p in CRC cells. Importantly, the inhibitory activities on CRC progression mediated by FTX deletion were reversed miR-192-5p down-regulation or EIF5A2 up-regulation.
Conclusion: LncRNA FTX functioned as an oncogene to contribute to CRC progression by regulating miR-192-5p/EIF5A2 axis, providing a novel insight into the pathogenesis of CRC and a promising therapeutic target for CRC treatment.
Keywords: FTX, miR-192-5p, EIF5A2, CRC, progression
Introduction
As the fourth most deadly cancer worldwide, there are almost 700,000 people died every year due to colorectal cancer (CRC).1 With the development in early diagnosis and effective therapeutic, the average survival times of advanced CRC have roughly doubled; however, the patients usually die within three years.1,2 Thus, extensive efforts to get deeper understanding underlying the molecular mechanisms pathogenesis of CRC are necessary for developing effective therapeutic strategies.
Long non-coding RNA (lncRNA) are a class of RNA molecules with longer than 200 bases in length and lack of protein-coding capacity. Emerging evidence has revealed that lncRNAs are implicated in diverse cellular and biological processes, including gene regulation at transcription or post-transcription level, chromatin dynamics and genomic imprinting.3,4 In recent years, aberrant expression of lncRNAs in many prevalent cancer types has been found, and alterations in the expression of lncRNAs are closely associated with tumorigenesis, metastasis, and tumor stage.5,6 LncRNAs can be novel excellent biomarkers and powerful therapeutic targets for cancer intervention.7 LncRNA five prime to Xist (FTX) is a kind of a highly conserved transcript with 2300 nucleotides encoded by FTX gene.8 It has been demonstrated that FTX functions as an oncogene to participate in the regulation of several cancers progressions, such as hepatocellular carcinoma, renal cell carcinoma and glioma.9–11 Besides that, FTX was also found to be elevated in CRC tissues and highly expressed FTX predicted poor overall survival, furthermore, overexpressed FTX significantly promoted the growth and progression of CRC through interaction with miR-215 and vimentin.12,13 However, the exact molecular mechanism underlying these effects in CRC remains vague.
MicroRNAs (miRNAs) are another type of non-coding RNAs that alter gene expressions in many processes, thereby regulating mRNA degradation or translational suppression.14 Up to date, many miRNAs have been recognized to play a critical role in the regulation of biological processes in various cancers, including CRC.15 Among these miRNAs, miR-192-5p (miR-192) was also identified to link with the progression of CRC and might be a promising target for CRC treatment.16 Recently, eukaryotic initiation factor 5A2 (EIF5A2), a novel candidate oncogene isolated from ovarian cancers,17 has also been observed to function as an oncogene to implicate in the regulation of CRC development and may serve as a promising target for molecular therapy.18,19 However, whether the miR-192-5p or EIF5A2, their detail regulatory networks in CRC are still unclear.
In this study, we aimed to investigate the expression pattern of FTX in CRC and to explore the regulatory effect of FTX on CRC progression as well as its underlying molecular mechanism in vivo and in vitro.
Materials and Methods
Tissue Specimens
Tumors and paracancerous tissues from 30 CRC patients were obtained from The Second Affiliated Hospital of Soochow University and were immediately preserved in liquid nitrogen until used. All CRC patients were histopathologically diagnosed by two independent pathologists, and the clinicopathological factors, including gender, age, tumor size, TNM stage, lymph node metastasis, and distant metastasis, in patients with CRC. Written informed consent had been collected from all patients, and this work was permitted by the Research Ethics Committee of The Second Affiliated Hospital of Soochow University.
Cell Culture and Transfection
The normal colon epithelial cell line FHC and CRC cell lines (HT-29, HCT-8, COLO 205, SW480 and LoVo) were obtained from American Type Culture Collection (APTT, Rockville, MD, USA) and were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) which supplemented with 10% fetal bovine serum (FBS; Gibco) at 37°C with 5% CO2.
The miRNA mimic or inhibitor targeting miR-192-5p (miR-192-5p or miR-192-5p inhibitor) and their corresponding negative control (NC mimic or NC inhibitor), short hairpin RNA (shRNA) targeting FTX (sh-FTX: CTGCTACGACACTGAATTC), shRNA scramble control (sh-NC), pcDNA3.1-EIF5A2 overexpression vector (EIF5A2), pcDNA3.1 empty vector (pcDNA) were synthesized by Genepharma (Shanghai, China). And then we used Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) to transfect these oligonucleotides or vectors into SW480 and LoVo cells.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Trizol reagent (Invitrogen) was utilized to isolate total RNAs from CRC tissue and cells following the standard introductions. For mRNA analysis, the Reverse Transcription System Kit (Takara, Dalian, China) was used to generate the complementary DNA (cDNA). For miRNA expression detection, cDNA was synthesized using the Takara RNA PCR kit (Takara). Next, SYBR Premix Ex Taq (Takara) was utilized for quantitative real-time PCR. The relative expression was calculated using 2−ΔΔCT methods with normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6. The primer sequences are presented as followed: miR-192-5p F, 5ʹ-GCGGCGGCTGACCTATGAATTG-3ʹand R, 5ʹ-ATCCAGTGCAGGGTCCGAGG-3ʹ; EIF5A2 F, 5ʹ-GGACGACCATGCAAAATAGTGG-3ʹand R, 5ʹ-TGCCCGTGAAAATATCAATTCCA-3ʹ; FTX F, 5ʹ-GTGTCTCTCTCTCTCTCTCTCTT-3ʹ and R, 5ʹ-CCTCTTCAGCAGTAGCATAGTT-3ʹ; GAPDH F, 5ʹ-ATTCCATGGCACCGTCAAGGCTGA-3ʹ and R, 5ʹ-TTCTCCATGGTGGTGAAGACGCCA-3ʹ;U6 F, 5ʹ-GCAGGAGGTCTTCACAGAGT-3ʹ and R, 5ʹ-TCTAGAGGAGAAGCTGGGGT-3ʹ.
Cell Proliferation
Transfected cells were seeded onto 96-well plates. After incubation overnight, each well was added with 10 μL of MTT solution ((3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma, St. Louis, MO) and incubated for another 4 h. At the end of incubation, each well was added with 150 μL DMSO (Sigma) after removing the supernatants. The absorbance value of each well was measured at 490 nm.
Cell Apoptosis
Annexin V-FITC/PI apoptosis detection kit (BD Biosciences, San Jose, CA, USA) was utilized to test cell apoptosis following the standard protocol after transfection. Briefly, resuspended cells were stained with 10 μL Annexin V-FITC and PI in the dark for 15 min. Subsequently, the apoptotic cells were analyzed by flow cytometry.
Transwell Assay
The abilities of migration and invasion of cells were analyzed using Transwell chambers with an 8-µm pore polycarbonate membrane (Corning Costar, Beijing, China) pre-coated without or with Matrigel (BD Biosciences). Briefly, transfected cells suspended in serum-free DMEM were seeded into the upper chamber Then 500 µL DMEM complete medium was added into the lower chamber as a chemoattractant. After 24 h incubation, cells on the low faces were fixed with 4% formaldehyde and stained with 0.1% crystal violet. Finally, migrated and invaded cells in five randomly selected visual fields were counted under a microscope.
Dual-Luciferase Reporter Assay
The FTX or EIF5A2 3ʹUTR containing the wild-type (WT) and mutated (MUT) sequences of miR-192-5p was synthesized and cloned into the pmirGLO vectors (Promega, Shanghai, China). Then, 10 μg constructed luciferase reporter vector combined with miR-192-5p mimics or NC mimics were transfected into SW480 and LoVo cells, respectively. Finally, a dual luciferase reporter assay kit (Promega) was applied to examine the relative luciferase activity.
Pull-Down Assay
Cells were transfected with biotin-labeled FTX (Bio-WT-FTX or Bio-MUT-FTX) and corresponding negative control (Bead), respectively. Then cells were lysed and the lysate was incubated with streptavidin magnetic beads to generate the RNA-RNA complexes. After elution, the level of miR-192-5p was analyzed by qRT-PCR.
Western Blot
Proteins were isolated from transfected cells using RIPA lysis buffer (Beyotime, Shanghai, China) and then were quantified with a bicinchoninic acid (BCA) method following the manufacturer’s instructions. An equal amount of the extracts were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA). After blocked with 5% non-fat milk for 1 h, the membrane was incubated with primary antibodies anti-EIF5A2 and anti-GAPDH, and followed by interaction with the HRP-conjugated secondary antibody. Finally, immunoreactive signals were visualized using Enhanced Chemiluminescence.
Xenograft Tumor Model
BALB/c nude mice (male, aged 4–6 weeks) were used for the xenograft assays. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. Experimental procedures were approved by the Institutional and Local Committee on the Care and Use of Animals of The Second Affiliated Hospital of Soochow University. All animals received humane care according to the National Institutes of Health (USA) guidelines. LoVo cells (2×106) transfected with sh-FTX or sh-NC were subcutaneously injected into the one flank of the nude mice. Subsequently, the tumor sizes were measured every seven days and the tumor volumes were calculated. After 30 days, all mice were killed and then tumor masses were weighted and used for subsequent molecular analysis.
Statistical Analysis
All statistical data from at least three times independently experiment was indicated as mean ± standard deviation (SD) and then was analyzed by GraphPad Prism 7 software (GraphPad Inc., San Diego, CA, USA). Student’s t-test or one-way analysis of variance (ANOVA) was performed to analyze the differences among multi groups. The correlation analysis was conducted using Pearson’s chi-square test. P < 0.05 exhibited a statistically significance.
Results
FTX Is Up-Regulated in CRC Tissues and Cell Lines
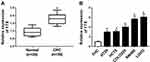
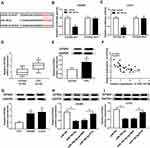
FTX expression was measured and results showed by contrast with the normal tissues and normal colon epithelial cell line FHC, FTX was significantly elevated in CRC tissues and cell lines, including HT-29, HCT-8, COLO 205, SW480 and LoVo (Figure 1A and B). Additionally, patients were grouped into a high FTX group and a low FTX group according to the median level, and we found higher FTX expression was correlated with tumor size, TNM stage, lymph Node metastasis and distant metastasis (P < 0.05, Table 1). Thus, these results suggested that FTX expression was dysregulated in CRC, and its high expression might be associated with the progression of CRC.
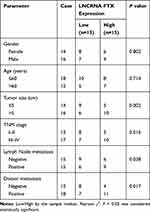 |
Table 1 Correlation Between FTX Expression and Clinicopathological Factors in Colorectal Cancer Patients |
FTX Deletion Inhibits Cell Malignant Properties in CRC in vitro
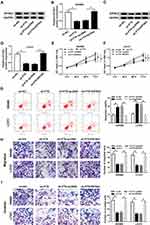
To investigate the potential biological function of FTX in CRC development, SW480 and LoVo cells were transfected with sh-FTX or sh-NC. Then an obvious down-regulation of FTX expression in cells transfected with sh-FTX was demonstrated (Figure 2A). Subsequently, MTT assay indicated knockdown of FTX inhibited cell proliferation in CRC (Figure 2B and C). Meanwhile, flow cytometry analysis showed FTX silence markedly induced apoptosis in SW480 and LoVo cells (Figure 2D). Furthermore, transwell assay implied decreased FTX significantly reduced the migration and invasion of SW480 and LoVo cells (Figure 2E and F). Besides that, FTX knockdown reduced the number of SW480 and LoVo cells in the S phase, while cells in the G0/G1 phase was accumulated, indicating cell cycle arrest (Figure S2). All the results suggested that FTX deletion could suppress CRC cell biological behavior in vitro.
FTX Is a Sponge of miR-192-5p in CRC Cells
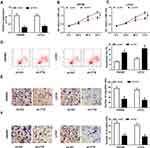
To explore the molecular mechanism underlying the promotion of FTX in CRC progression, the downstream targeted miRNAs were predicted using starBase v2.0 software, and miR-192-5p was predicted that might be a target of FTX with a putative binding site (Figure 3A). To confirm this prediction, Dual-luciferase reporter assay was conducted and we discovered that miR-192-5p mimic transfection obviously reduced the luciferase activity of WT-FTX reporter but not MUT-FTX reporter in SW480 and LoVo cells (Figure 3B and C). Moreover, FTX pull-down in SW480 and LoVo cells was significantly enriched for miR-192-5p compared with the empty vector (Beads) and mutant type FTX (Figure 3D and E). Subsequently, the expression of miR-192-5p in CRC tumors and corresponding non-tumor tissues was detected using qRT-PCR assay and CRC tissues expressed lower miR-192-5p levels than non-tumor tissue (Figure 3F). Meanwhile, a negative correlation between FTX and miR-192-5p expression was observed in CRC tissues (Figure 3G). Additionally, miR-192-5p was also decreased in cell lines (Figure 3H). Besides that, FTX deletion was found to induce miR-192-5p expression and miR-192-5p mimic transfection negatively affected FTX levels (Figure 3I and J). Based on the results, we verified that FTX directly interacted with miR-192-5p and negatively regulated its expression in CRC cells.
FTX Deletion Inhibits Cell Malignant Properties in CRC by Sponging miR-192-5p in vitro
As shown in Figure S1, miR-192-5p overexpression inhibited the migration and invasion of SW480 and LoVo cells (Figure S1A and B). Thus, whether miR-192-5p involved in the FTX mediated promotion on CRC progression was elucidated, SW480 and LoVo cells were transfected with sh-NC, sh-FTX, sh-FTX + miR-192-5p inhibitor, sh-FTX + NC inhibitor. Afterwards, we found decreased miR-192-5p could reverse FTX deletion induced inhibition on proliferation (Figure 4A and B), migration and invasion (Figure 4E and F), enhancement on apoptosis (Figure 4C and D) in SW480 and LoVo cells. These data suggested that FTX deletion regulated CRC progression in by sponging miR-192-5p in vitro.
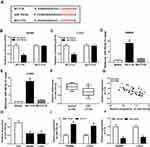
EIF5A2 Is a Target of miR-192-5p in CRC Cells
According to the prediction of starBase2.0 program, EIF5A2 might be a target of miR-192-5p with a putative binding site in CRC cells (Figure 5A). Immediately, Dual-luciferase reporter assay results displayed that miR-192-5p mimic transfection significantly reduced the luciferase activity of EIF5A2-WT reporter, while there was no change in EIF5A2-MUT reporter in SW480 and LoVo cells (Figure 5B and C), indicating the direct interaction between miR-192-5p and EIF5A2. After that, the mRNA and protein expression of EIF5A2 was detected and a markedly increased EIF5A2 level was demonstrated in CRC tissues compared with that in normal tissues (Figure 5D and E), besides, a negative correlation between miR-192-5p and EIF5A2 expression was also investigated in CRC tissues (Figure 5F). Moreover, EIF5A2 was also elevated in CRC cell lines, which consistent with the results in CRC tissues (Figure 5G). Taken together, miR-192-5p targetedly repressed EIF5A2 expression in CRC cells. After that, expression analysis was performed and we found the expression of EIF5A2 was notably suppressed by miR-192-5p mimic transfection, which could be restored by overexpressed FTX in SW480 and LoVo cells (Figure 5H and I), indicating FTX could regulate EIF5A2 by directly interacting with miR-192-5p in SW480 and LoVo cells.
FTX Silence Suppresses Cell Biological Behavior in CRC Through Regulating EIF5A2 in vitro
In this study, we discovered that EIF5A2 up-regulation promoted SW480 and LoVo cell migration and invasion (Figure S1C and D). Besides, considering that FTX regulated EIF5A2 via miR-192-5p, we supposed that EIF5A2 might be implicated in FTX mediated regulation in CRC cells. To verify this hypothesis, SW480 and LoVo cells were transfected with sh-NC, sh-FTX, sh-FTX + pcDNA, sh-FTX + EIF5A2. Then we found both the mRNA and protein expression of EIF5A2 was inhibited by FTX down-regulation, while was restored by EIF5A2 up-regulation in SW480 and LoVo cells (Figure 6A–D). Afterwards, rescue assay implied that EIF5A2 overexpression antagonized FTX deletion-mediated suppression on proliferation (Figure 6E and F), migration and invasion (Figure 6H and I), as well as stimulation on apoptosis (Figure 6G) in SW480 and LoVo cells. Thus, we illustrated that FTX silence suppressed CRC progression through regulating EIF5A2 in vitro.
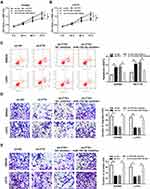
Knockdown of FTX Inhibits Tumor Growth of CRC in vivo
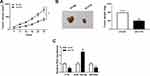
To further investigate the effect of FTX in animals, LoVo cells stably transfected with sh-FTX was used to establish xenograft models and then we found that tumor volume and weight were greatly suppressed in sh-FTX group compared with these in sh-NC group (Figure 7A and B). Furthermore, molecular analysis was performed in harvested tumor tissues and results showed the expression of FTX and EIF5A2 was obviously decreased while miR-192-5p level was increased in sh-FTX group compared with that in sh-NC group (Figure 7C). Therefore, we demonstrated that FTX deletion might suppress tumor growth in vivo by regulating miR-192-5p and EIF5A2 expression.
Discussion
Increasing researches have reported that lncRNAs involve in various aspects of cell biology and potentially regulate tumor development.20 In CRC, numerous lncRNAs have been identified to implicate in the progression of CRC by stimulating or inhibiting diverse biological processes. For example, lncRNA CRNDE promoted the proliferation and chemoresistance of CRC cells by interacting with miR-181a-5p and activating the Wnt/β-catenin signaling pathway.21 Chen et al found that lncRNA XIST expedited CRC progression and metastasis by acting as competing for endogenous RNA (ceRNA) for miR-200b-3p to regulate ZEB1 expression.22 LncRNA-UCA1 was found to accelerate cell proliferation and 5-fluorouracil resistance in CRC via directly sponging miR-204-5p.23 All the evidence indicated the important regulatory role of lncRNAs in CRC progression.
FTX was reported to be a novel cancer-related lncRNA in different tumor types. For instance, FTX was elevated in gliomas and promoted glioma cells progression by a negative interaction with miR-342-3p.10 Li et al indicated FTX induced aerobic glycolysis by activating the PPARγ pathway, thereby promoting hepatocellular carcinoma progression.9 Besides, FTX was also implicated in the development of CRC.24 Nevertheless, the molecular mechanism of FTX in CRC is still unclear. In the current study, the level of FTX was found to be up-regulated in CRC tissues and cell lines, implying the important role of FTX in CRC development. Subsequently, functional experiments were performed and we found FTX deletion suppressed CRC cell proliferation, migration and invasion, involved in the induction of apoptosis in vitro. Besides that, using shRNAs against FTX could dramatically repress CRC growth in vivo.
An increasing number of researches have proposed the widespread existence of the interaction among lncRNA-miRNA-mRNA networks involving ceRNAs, in which the lncRNA can compete for miRNA binding to regulate the targeted mRNA expression.25 For example, overexpressed HOXD-AS1 competitively bound to miR-130a-3p to prevent SOX4 degradation, thereby facilitating hepatocellular carcinoma metastasis.26 LncRNA H19 regulated the expression of Vimentin, ZEB1, and ZEB2 genes implicated in EMT via functioning as a ceRNA for miR-138 and miR-200a, which promoted EMT progression and accelerated tumor growth in CRC.27 In this study, using the bioinformatics databases, miR-192-5p was identified that might interact with FTX, and then this prediction was confirmed by Dual-luciferase reporter and pull-down assays; besides that, expression analysis indicated FTX deletion induced miR-192-5p expression and miR-192-5p mimic transfection negatively affected FTX levels, which suggests the regulating relationship between FTX and miR-192-5p. After that, rescue assay was conducted and FTX could inhibit CRC progression by sponging miR-192-5p in CRC cells.
EIF5A is a translation factor that affects both initiation and elongation and involved in transcription, mRNA turnover, and nucleocytoplasmic transport. EIF5A2, a second isoform of EIF5A, has been observed to be up-regulated in many type tumors and is an important regulator in the tumorigenesis of cancers.28,29 Importantly, the aberrant expression has also been investigated in CRC. EIF5A2 was associated with epithelial mesenchymal transition (EMT),19 and Bao et al found EIF5A2 was elevated in CRC, and EIF5A2 down-regulation enhanced the chemosensitivity to doxorubicin in colon cancer cells by inhibiting cell EMT.30 Zhu et al demonstrated that EIF5A2 was an independent predictor of shortened survival and promoted CRC cell invasiveness and inhibited EMT by up-regulating metastasis-associated protein 1 (MTA1) partly dependent on C-myc.31 Thus, EIF5A2 could be used as a novel prognostic marker and/or effective therapeutic target for CRC. In this study, EIF5A2 was predicted and confirmed to be a target of miR-192-5p. EIF5A2 was up-regulated in CRC tissues and cell lines, and co-transfection analysis showed that FTX functioned as a ceRNA for miR-192-5p to modulate EIF5A2 expression in CRC cells. Besides that, the inhibitory effects on CRC progression mediated by FTX silence could be reversed by EIF5A2 overexpression.
In conclusion, our results showed FTX was increased in CRC tissues and cell lines, and high FTX expression promoted CRC cell malignant properties in vitro and tumor growth in vivo. Also, we also identified the FTX/miR-192-5p/EIF5A2 regulatory network in the development of CRC, indicating a potential therapeutic target for the treatment of CRC.
Disclosure
The authors declare that they have no financial conflicts of interest in this work.
References
1. Brody H. Colorectal cancer. Nature. 2015;521(7551):S1. doi:10.1038/521S1a
2. Day LW, Velayos F. Colorectal cancer screening and surveillance in the elderly: updates and controversies. Gut Liver. 2015;9(2):143. doi:10.5009/gnl14302
3. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
4. Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:93–95. doi:10.1016/j.steroids.2017.12.012
5. Vitiello M, Tuccoli A, Poliseno L. Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol. 2015;38(1):17–28. doi:10.1007/s13402-014-0180-x
6. Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15(1):43. doi:10.1186/s12943-016-0530-6
7. Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474(24):4219–4251. doi:10.1042/BCJ20170079
8. Chureau C, Chantalat S, Romito A, et al. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet. 2011;20(4):705–718. doi:10.1093/hmg/ddq516
9. Li X, Zhao Q, Qi J, et al. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARgamma pathway in hepatocellular carcinoma. Int J Oncol. 2018;53(2):551–566. doi:10.3892/ijo.2018.4418
10. Zhang W, Bi Y, Li J, et al. Long noncoding RNA FTX is upregulated in gliomas and promotes proliferation and invasion of glioma cells by negatively regulating miR-342-3p. Lab Invest. 2017;97(4):447–457. doi:10.1038/labinvest.2016.152
11. He X, Sun F, Guo F, et al. Knockdown of long noncoding RNA FTX inhibits proliferation, migration, and invasion in renal cell carcinoma cells. Oncol Res. 2017;25(2):157–166. doi:10.3727/096504016X14719078133203
12. Yang Y, Zhang J, Chen X, et al. LncRNA FTX sponges miR-215 and inhibits phosphorylation of vimentin for promoting colorectal cancer progression. Gene Ther. 2018;1.
13. Guo X-B, Hua Z, Li C, et al. Biological significance of long non-coding RNA FTX expression in human colorectal cancer. Int J Clin Exp Med. 2015;8(9):15591.
14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
15. Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10(3):197–214. doi:10.1242/dmm.027441
16. Zhao H, Chen J, Chen J, et al. miR‐192/215‐5p act as tumor suppressors and link crohn’s disease and colorectal cancer by targeting common metabolic pathways: an integrated informatics analysis and experimental study. J Cell Physiol. 2019. doi:10.1002/jcp.28709
17. Guan XY, Sham JS, Tang TC, et al. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res. 2001;61(9):3806–3809.
18. Deng B, Wang B, Fang J, et al. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci Rep. 2016;6(1):28301. doi:10.1038/srep28301
19. Kolligs FT. An alternative way for epithelial-to-mesenchymal transition in colorectal cancer via EIF5A2? Gut. 2012;61(4):473–474. doi:10.1136/gutjnl-2011-301091
20. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi:10.1158/2159-8290.CD-11-0209
21. Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16(1):9. doi:10.1186/s12943-017-0583-1
22. Chen DL, Chen LZ, Lu YX, et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8(8):e3011. doi:10.1038/cddis.2017.421
23. Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6(1):23892. doi:10.1038/srep23892
24. Iwaya T, Sato K, Kume K, et al. Overexpression of Long Noncoding RNA FTX Was Associated with Colorectal Cancer Progression. AACR; 2015.
25. Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
26. Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi:10.1186/s12943-017-0680-1
27. Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525. doi:10.18632/oncotarget.4154
28. Mathews MB, Hershey JW. The translation factor eIF5A and human cancer. Biochim Biophys Acta. 2015;1849(7):836–844. doi:10.1016/j.bbagrm.2015.05.002
29. Rossi D, Kuroshu R, Zanelli CF, et al. eIF5A and EF-P: two unique translation factors are now traveling the same road. Wiley Interdiscip Rev RNA. 2014;5(2):209–222. doi:10.1002/wrna.1211
30. Bao Y, Lu Y, Wang X, et al. Eukaryotic translation initiation factor 5A2 (eIF5A2) regulates chemoresistance in colorectal cancer through epithelial mesenchymal transition. Cancer Cell Int. 2015;15(1):109. doi:10.1186/s12935-015-0250-9
31. Zhu W, Cai M-Y, Tong Z-T, et al. Overexpression of EIF5A2 promotes colorectal carcinoma cell aggressiveness by upregulating MTA1 through C-myc to induce epithelial–mesenchymaltransition. Gut. 2012;61(4):562–575. doi:10.1136/gutjnl-2011-300207
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.