Back to Journals » Cancer Management and Research » Volume 13
LncRNA FOXD3-AS1 Promotes Tumorigenesis of Glioma via Targeting miR-128-3p/SZRD1 Axis
Authors Li Z, Li M, Xia P, Wang L, Lu Z
Received 10 July 2021
Accepted for publication 16 November 2021
Published 7 December 2021 Volume 2021:13 Pages 9037—9048
DOI https://doi.org/10.2147/CMAR.S324920
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
This paper has been retracted.
Zhang Li, Ming Li, Pengcheng Xia, Lili Wang, Zhiming Lu
Department of Clinical Laboratory, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, 250012, People's Republic of China
Correspondence: Zhiming Lu
Department of Clinical Laboratory, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, 250012, People's Republic of China
Tel +86-13658608997
Email [email protected]
Background: The aim of the current study was to investigate the roles of LncRNA FOXD3-AS1 (FOXD3-AS1) in the glioma progression, and its underlying mechanism of competing endogenous RNA (ceRNA) network of FOXD3-AS1/miR-128-3p/SZRD1.
Materials and Methods: The FOXD3-AS1 expression and its prognostic relation were detected by bioinformatics tool. Next, glioma cell lines (HS683, U251, T98G, and SNB-19) were used to verify the FOXD3-AS1 expression. Furthermore, the roles of the FOXD3-AS1/miR-128-3p/SZRD1 axis on the glioma development in vitro and in vivo were examined.
Results: Bioinformatics analysis showed that FOXD3-AS1 was upregulated in the glioma and linked with poor prognosis. Consistently, FOXD3-AS1 level was overexpressed in the glioma cell lines (HS683 and U251). Subsequently, we verified that silencing of FOXD3-AS1 (si-FOXD3-AS1) restrained the cell proliferation, invasion, and tumor growth in vivo, and induced G0/G1 arrest, and promoted apoptosis. Further study also stated that FOXD3-AS1 interacted with miR-128-3p and SZRD1 was the target gene of miR-128-3p. Moreover, overexpression of miR-128-3p restrained the cell proliferation and metastasis of glioma, and reduced the SZRD1 level. Rescue assay illustrated that miR-128-3p inhibitor could reverse the suppressive impact of si-FOXD3-AS1 on the glioma progression. Similarly, SZRD1 overexpression could neutralize the influences of miR-128-3p mimic on glioma progression.
Conclusion: FOXD3-AS1 promoted the tumorigenesis of glioma, and exerted its function to modulate SZRD1 by targeting miR-128-3p.
Keywords: glioma, LncRNA FOXD3-AS1, MiR-128-3p, SZRD1, proliferation, invasion
Introduction
As a dominating primary intracranial tumor, glioma is featured by high aggression, poor prognosis, and high lethality.1 On the basis of the grading criteria, glioma is classified as low-grade glioma (LGG, I–II) and high-grade glioma (HGG, III–IV).2 Of these, patients with LGG have longer overall survival in contrast to HGG.3 Glioblastomas (GBM), refers to the most aggressive type (grade IV), with a proportion of 65% in brain tumors.4 Despite advances in therapeutic treatment, most patients achieve unsatisfactory efficacy and have poor prognosis, especially with GBM.5 Like other tumors, the progression of glioma is a complex procedure involving changes of pathology, genes, and pathways.6 Currently, more attention has been paid to the molecular mechanism of glioma progression.7 However, there is no definite conclusion about pathogenesis of glioma. Thus, elucidating the molecular mechanism is an urgent need for glioma therapy.
Accumulating evidence has stated that LncRNAs exert crucial roles on the progression of various cancers including glioma.8–10 Moreover, a previous study has reported that LncRNAs as competing endogenous RNAs (ceRNAs) by sponging miRNAs to directly target the mRNAs is the key mechanism for regulation of tumor growth and metastasis.11 In terms of glioma, recent studies have verified this point.12,13 Those findings suggested that the LncRNA as ceRNA may be the underlying mechanism to glioma development.
LncRNA FOXD3-AS1 (FOXD3-AS1) is highly expressed and may exert the crucial function on cancer progression through the ceRNA network, including lung cancer, malignant melanoma, and colon adenocarcinoma.14–16 Consistently, FOXD3-AS1 is upregulated in glioma, which has been demonstrated in clinical specimen detection.17 However, no reports of FOXD3-AS1 as ceRNA regulating glioma development are available. Microarray studies have been conducted to screen the aberrantly expressed miRNAs in glioma and found that miR-128-3p is observably declined.18,19 Moreover, miR-128-3p is the target gene of FOXD3-AS1, which has been elucidated in cervical cancer.20 Another study has also demonstrated that miR-128-3p suppresses the proliferation and metastasis of glioma cells via binding to 3ʹ-UTR of SZRD1, thereby influencing the glioma tumorigenicity.21 Thus, we speculated that FOXD3-AS1 regulating miR-128-3p/SZRD1 axis may be an underlying mechanism in glioma progression. In the current study, we first verified that functioned as ceRNA to regulate the glioma progression. In addition, miR-128-3p/SZRD1 axis as a novel target of FOXD3-AS1 in regulation of glioma development may supply theoretical foundation for further study.
Materials and Methods
Cell Culture
Glioma cell lines (HS683, U251, T98G, and SNB-19) and human normal astrocyte (HEB) were purchased from BeNa Culture Collection (Beijing, China), and cultured in DMEM (BI, Israel) supplemented with 10% FBS (Solarbio, Beijing, China), and 100 U/mL penicillin and 100 mg/mL streptomycin (Meilunbio, Dalian, China).
Cell Transfection
After reaching 80% confluence, HS683 and U251 cells were transfected with siRNA-FOXD3-AS1 (si-FOXD3-AS1) and negative control (si-NC) (Genechem, Shanghai, China) respectively according to the Lipofectamine 2000 protocol. Similarly, pcDNA-SZRD1 (OE-SZRD1) and empty vector was synthesized by Genechem (Shanghai, China). The miR-128-3p inhibitor, miR-128-3p mimic and its negative control were obtained from RiboBio (Guangzhou, China). Cells in the control group were untreated with anything. After being cultured for 48 h, cells were harvested.
Quantitative Real-Time PCR Assay
Total RNA was extracted by TRIzol reagent (CWBio, Beijing, China). The first strand cDNA for FOXD3-AS1 and SZRD1 was synthesized by cDNA Synthesis kit (Accurate Biology, Changsha, China). Next, PCR reaction was performed on CFX connect detection system (Bio-Rad, Hercules, CA, USA). GAPDH was used as the internal control for quantification of FOXD3-AS1 and SZRD1 levels, and U6 for miR-128-3p level, respectively. Primers were listed as the following: FOXD3-AS1 forward, 5ʹ- ACCAGAGGAAGGAGCACGA-3ʹ, reverse, 5ʹ-AGAAGCACCACTGTCCATCC-3ʹ; miR-128-3p forward, 5ʹ-TCACAGTGAACCGGTC-3ʹ, and reverse, 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ; SZRD1 forward, 5ʹ-ATGAGGAGGTCGCTGAGAG-3ʹ, and reverse, 5ʹ-GGAAGGCTATCGTCCTGAATC-3ʹ; GAPDH forward, 5ʹ-AGAAGGCTGGGGCTCATTTG-3ʹ, reverse, 5ʹ-AGGGGCCATCCACAGTCTTC-3ʹ; U6 forward, 5ʹ- GCTCGCTTCGGCAGCACA-3ʹ, and reverse, 5ʹ- GAACGCTTCACGAATTTGCGTG-3ʹ. The relative expression was quantified by 2–ΔΔCT method.
Western Blotting Assay
Protein was extracted from HS683 and U251 cells using RIPA lysis buffer containing protease inhibitor (Bosterbio, Wuhan, China). Protein was separated and then transferred onto PVDF membranes. After blockage with 5% skimmed milk, the membranes were incubated with anti-SZRD1 (1:3000, ab95957, Abcam, MA, USA), and anti-GAPDH (1:5000, 60004-1-Ig, Proteintech), followed by HRP-conjugated secondary antibody (1:5000, ZB-2301, ZSGB). The protein blots were visualized using the enhanced chemiluminescence (ECL) kit and captured using chemiluminescent imaging system (Tanon, Shanghai, China). GAPDH was used as internal control.
3-(4, 5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
After transfection, HS683 and U251 cells were cultured (2×103 cells/well) for 24 h, 48 h, 72 h, and 96 h. Afterwards, MTT with concentration of 5 mg/mL was added for incubation of four hours. The absorbance at 490 nm was detected by spectrophotometer (Multiskan Sky, Thermo Fisher, Waltham, MA, USA).
Flow Cytometry Assay
For analysis of cell apoptosis, HS683 and U251 cells (1×106 cells/mL) were obtained after 48 h of transfection, and washed with cool phosphate buffered saline (PBS) three times. Cell apoptosis was detected by flow cytometer (NovoCyte, Agilent, Palo Alto, CA, USA) with annexin V-propidium iodide (PI) kit (Meilunbio, Dalian, China). To detect cell cycle, collected cells were fixed in the 75% ethanol. Having been washed, the cells were stained with PI, followed by analysis with flow cytometer (NovoCyte, Agilent, Palo Alto, CA, USA).
Transwell Assay
HS683 and U251 cells (1×105 cells/mL) were placed in a transwell chamber (Corning, Lowell, MA, USA), and the culture medium was in the lower chamber. Subsequently, the invasion and migration cells were fixed with 4% paraformaldehyde and then dyed with 0.1% crystal violet. The number of cells was quantified by microscope (CKX53, Olympus, Tokyo, Japan).
Dual-Luciferase Reporter Assay
The wild type (WT) and mutant (MU) fragments of FOXD3-AS1 were cloned into the pmirGLO vector to establish the recombinant vectors (pmirGLO-FOXD3-AS1-WT and pmirGLO-FOXD3-AS1-MU). Consistently, WT and MU fragments of SZRD1 were cloned into the abovementioned vector. Subsequently, 8 μg/mL pmirGLO vector and 16 μg/mL miR-128-3p mimic or mimics-NC were co-transfected into U251 cells. After transfection for 48 h, luciferase activities were detected.
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted as previously described.22 Briefly, transfected HS683 and U251 cells were lysed and incubated with magnetic beads conjugated with human anti-Argonaute2 (Ago2) antibody, negative control (anti-IgG) or input Control (Millipore) for six hours. Subsequently, immunoprecipitated RNA was isolated and detected by qRT-PCR.
Tumorigenesis Assay in vivo
A total of 72 nude mice (five weeks old) were obtained from Hangzhou Ziyuan Laboratory nimal Science and Technology Co. Ltd (Hangzhou, China). The mice were divided into six groups, which were inoculated with HS683 and U251 cells (1×106 cells/mL) transfected with control, sh-FOXD3-AS1, sh-NC; control, miR-128-3p mimic, and mimics-NC, respectively (12 mice per group). The nude mice in the control group were injected with glioma cells untreated. The tumor volume was assessed from one week to six weeks after tumor inoculation (once every week). Subsequently, mice were sacrificed by cervical dislocation and the tumor tissues were collected for the further experiments.
Bioinformatics Analysis
The FOXD3-AS1 expression profiles (GSE147352) in glioma patients were retrieved from the Gene Expression Omnibus (GEO) database. The dataset included 85 GBM samples, 18 LGG samples and 15 normal samples. Gene expression was estimated using fragments per kilobase of transcript per million mapped reads (FPKM) value and then Boxplot was visualized using the ggplot2 package in R Language. The differential expression between normal and tumor groups was analyzed by R package Deseq2 on basis of following criteria: fold change >2 and Padj <0.05. Samples were collected according to inclusion criteria as following: (1) patients were diagnosed as glioma; (2) data were available on expression of FOXD3-AS1; and (3) data of samples on FOXD3-AS1 expression had corresponding control group. Samples were excluded according to following exclusion criteria: (1) FOXD3-AS1 expression was not included; (2) control group was not included; and (3) cell samples were used.
The Cancer Genome Atlas (TCGA) data were applied to analyze the relationship between FOXD3-AS1 expression and patient’s prognosis. 638 glioma patients with FOXD3-AS1 expression and survival information were included into current analysis. The survival analysis was performed using R language with “Survival” package and survival curves were plotted using “Survminer” package. Samples were collected according to inclusion criteria as following: (1) patients were diagnosed as glioma including LGG and GBM; (2) data were available on expression of FOXD3-AS1; and (3) data of samples on FOXD3-AS1 expression had corresponding survival information. Samples were excluded according to following exclusion criteria: (1) survival information was not included; and (2) patients were diagnosed as other types of cancer except glioma. The target bites between miRNA and lncRNA (miRNA and mRNA) were predicted by StarBase v3.0.
Statistical Analysis
Data were presented as the means ± standard deviation (SD) and analyzed by SPSS 20.0 statistical software. The differences in the multiple groups were compared by one-way ANOVA followed by LSD test, respectively. P<0.05 was termed as statistically significant.
Results
FOXD3-AS1 Was Upregulated in Glioma and Correlated with Poor Prognosis
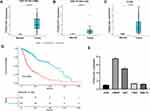
To investigate the function of FOXD3-AS1 in glioma, we downloaded the FOXD3-AS1 expression profiles (GSE147352) in glioma patients from GEO database. As displayed in Figure 1A and B, the FOXD3-AS1 expression in the GBM and LGG tissues (tumor group) was higher than that in normal tissues (normal group) (P<0.01) (Figure 1A and B). Consistently, data from TCGA database showed that FOXD3-AS1 in the tumor group including GBM and LGG was higher than that in the normal group (P<0.01) (Figure 1C). In addition, it also found that FOXD3-AS1 expression in glioma patients including GBM and LGG was negatively correlated with overall survival (P<0.01) (Figure 1D). Similarly, FOXD3-AS1 expression in glioma cell lines (HS683 and U251) was higher than that of HEB (Figure 1E) (P<0.01), indicating that HS683 and U251 cell lines can be used for further study.
FOXD3-AS1 Sponged miR-128-3p in Glioma
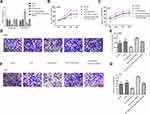
As shown in Figure 2A, complementary binding sites were observed between FOXD3-AS1 and miR-128-3p. Furthermore, results of RIP assay showed the enrichment of FOXD3-AS1 and miR-128-3p in the anti-Ago2 group compared with anti-IgG group (P<0.01) (Figure 2B and C). Luciferase reporter assay stated that overexpression of miR-128-3p significantly reduced the luciferase activity of FOXD3-AS1-WT (P<0.01) (Figure 2D). Thus, the aforementioned findings indicated that FOXD3-AS1 could directly regulate the miR-128-3p.
Silencing of FOXD3-AS1 Suppressed Proliferation and Invasion of Glioma Cells via Modulating miR-128-3p
To examine the roles of FOXD3-AS1 on glioma, silencing of FOXD3-AS1 (si-FOXD3-AS1) was applied in HS683 and U251 cells. qRT-PCR results revealed FOXD3-AS1 was notably decreased, but miR-128-3p expression was prominently increased in the si-FOXD3-AS1 group, in comparison with si-NC and control groups (P<0.01) (Figure 3A). Following, MTT results showed that cell viability in the si-FOXD3-AS1 group was notably reduced after 48 h, however, inhibition of miR-128-3p elevated the cell viability after 48 h compared with si-NC and control groups (P<0.05) (Figure 3B and C). Similarly, si-FOXD3-AS1 suppressed the invasion abilities, while inhibition of miR-128-3p rescued the roles of si-FOXD3-AS1 on invasion of HS683 and U251 cells (P<0.01) (Figure 3D–G). No conspicuous difference was observed among the si-FOXD3-AS1+miR-128-3p inhibitor, si-NC, and Control groups (P>0.05).
Knockdown of FOXD3-AS1 Influenced Cell Cycle and Promoted Apoptosis of Glioma Cells via Modulating miR-128-3p
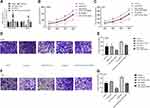
As shown in Figure 4A–D, the cell proportion at G0/G1 phase was prominently increased in the si-FOXD3-AS1 group, however miR-128-3p inhibitor reduced the cell proportion at G0/G1 phase (P<0.01). The cell proportion at different phases in si-FOXD3-AS1+miR-128-3p inhibitor group exhibited no obvious difference in contrast to control and si-NC groups (P>0.05). In the aspect of cell apoptosis, we discovered that apoptosis rate was notably increased in the si-FOXD3-AS1 group, whereas it was decreased in the miR-128-3p inhibitor group in contrast to control and si-NC groups (P<0.01). Moreover, miR-128-3p inhibitor can reverse the influences of si-FOXD3-AS1 on apoptosis rate (P<0.01) (Figure 4E–H). Collectively, the aforementioned results concluded that si-FOXD3-AS1 influenced cell cycle and facilitated apoptosis of glioma cells via modulating miR-128-3p.
Silencing of FOXD3-AS1 Inhibited Tumorigenesis in vivo via Modulating miR-128-3p
To validate the effect of FOXD3-AS1 on glioma in vitro, xenograft tumor model was conducted. First, the transfection efficacy was shown in Figure 5A, FOXD3-AS1 expression was decreased while miR-128-3p level increased after transfected with sh-FOXD3-AS1 (P<0.01). Subsequently, we found that the tumor size and weight of the sh-FOXD3-AS1 group was remarkably lessened in contrast to control and sh-NC groups (P<0.05) (Figure 5B–G).
MiR-128-3p Targeted the 3ʹ-UTR of SZRD1
As shown in Figure 6A, the 3ʹ-UTR of SZRD1 contains a targeting site for the region of miR-128-3p. Luciferase reporter assays showed that the relative luciferase activity of SZRD1-3ʹ-UTR-WT was remarkably decreased in overexpressed miR-128-3p group (Figure 6B).
Western blotting indicated that si-FOXD3-AS1 suppressed the SZRD1 expression, while miR-128-3p inhibitor elevated the SZRD1 level in comparison with control and si-NC groups (P<0.01). Surprisingly, SZRD1 level in the si-FOXD3-AS1+miR-128-3p inhibitor group did not differ from control and si-NC groups (P>0.05) (Figure 6C–F). Collectively, SZRD1 is the target gene of miR-128-3p regulated by FOXD3-AS1.
Overexpressed miR-128-3p Suppressed Proliferation and Invasion of Glioma Cells via Modulating SZRD1
MiR-128-3p mimic and OE-SZRD1 were co-transfected into HS683 and U251 cells, and it turned out that miR-128-3p expression was remarkably elevated in the miR-128-3p mimic group while SZRD1 was decreased (P<0.01). Besides, OE-SZRD1 reversed the influence of miR-128-3p in the aspect of SZRD1 level (Figure 7A). MTT results showed that cell viability was lowered after transfection with miR-128-3p mimic, whereas increased after transfection with OE-SZDR1 (P<0.05). There was no distinct difference in the OD490 value among the miR-128-3p mimic+OE-SZRD1, mimics-NC and control groups (P>0.05) (Figure 7B and C). Consistently, the number of invasion cells was reduced in miR-128-3p mimic group compared with mimics-NC and control groups (P<0.01). Nonetheless, transfection with OE-SZRD1 enhanced the invasion ability of miR-128-3p mimic (P<0.01) (Figure 7D–G).
Overexpression of miR-128-3p Inhibited Tumorigenesis in vivo via Modulating SZRD1
As shown in Figure 8A, qRT-PCR results validated transfection of miR-128-3p mimic (P<0.01). In vivo, it indicated that tumor size and weight of miR-128-3p mimic group was conspicuously decreased compared with control and mimics-NC groups (P<0.01) (Figure 8B–G). Importantly, the schematic for the regulatory relationship among FOXD3-AS1, miR-128-3p, and SZRD1 in glioma was shown in Figure 9. Those finding proved that overexpression of miR-128-3p restrained tumorigenesis in vivo via modulating SZRD1.
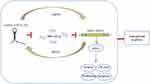 |
Figure 9 Schematic for the regulatory relationship among FOXD3-AS1, miR-128-3p, and SZRD1 in glioma. |
Discussion
To our knowledge, dysregulation of LncRNAs frequently occurs in the various cancers, which is deemed as a master regulator for disease progression.23 Here, our study showed that FOXD3-AS1 was upregulated in glioma and correlated with poor prognosis using bioinformatic analysis. In addition, FOXD3-AS1 level in the glioma cell lines was consistent with bioinformatic analysis. Furthermore, knockdown of FOXD3-AS1 suppressed the tumorigenesis of glioma via targeting miR-128-3p/SZRD1 axis, as evidenced by cell viability, invasive ability, apoptosis rate, cell cycle, and tumorigenesis in vivo.
FOXD3-AS1, is a 963-bp lncRNA, which is located in the chromosome 1p31.3 upstream of FOXD3 promoter,24 indicating that its function is closely associated with adjacent protein-coding transcripts.25 Beyond that, FOXD3-AS1 acts as independent prognostic indicator for prediction of neuroblastoma progression, which has been proved.24 Similarly, dysregulation of FOXD3-AS1 expression is observed in numerous cancers.14,26,27 In terms of glioma, Chen et al17 have clarified that si-FOXD3-AS1 inhibited the tumor development as demonstrated by cell proliferation, migration, and invasion. Consistently, our study found that FOXD3-AS1 was highly expressed in glioma, and si-FOXD3-AS1 restrained the tumor growth in vivo and in vitro. In addition, glioma patients with overexpressed FOXD3-AS1 had lower overall survival, indicating FOXD3-AS1 was closely associated with poor prognosis. Furthermore, Wang et al28 have demonstrated that FOXD3-AS1 may be functioning as ceRNA to exert the pivotal role in the regulation of cancer progression. To date, knowledge about FOXD3-AS1 ceRNA network involved in glioma progression remains vague.
MiR-128-3p was initially discovered in GBM,29 which attracts more attention at present because it acts as a tumor suppressor and early diagnostic indicator.30,31 A similar study has verified that miR-128-3p can increase the sensitivity of chemotherapy of colorectal cancer.32 In the gliomas, Bendahou et al33 have revealed that patients with high miR-128-3p expression have longer overall survival than low miR-128-3p using bioinformatic analysis. A similar study has demonstrated that miR-128 may be effective therapy for disruption of tumor-relevant phenotypes and tumor ingrowth.34 Furthermore, a previous study has reported that miR-128-3p restrained the cell proliferation and accelerated apoptosis.18 Aforementioned findings were aligned with our results. Our study showed that overexpression of miR-128-3p restrained proliferation and tumorigenesis in vivo, and accelerated apoptosis of glioma cells. On the contrary, inhibition of miR-128-3p accelerated the glioma development. In regard to the interaction between miRNAs and LncRNAs, previous studies have been reported. For example, a previous study reported by Fu et al22 has uncovered that miR-128-3p recuses the regulation of LncRNA PVT1 on glioma tumorigenesis. Another study also showed that inhibition of miR-128-3p eliminates the influence of si-LINC00346 on metastasis of glioma.21 However, the relationship between miR-128-3p and FOXD3-AS1 in glioma has not been reported. Accordingly, our study firstly disclosed that miR-128-3p inhibitor reversed the effects of si-FOXD3-AS1 on glioma progression. Taken together, FOXD3-AS1 may be involved in the modulation of glioma development via sponging miR-128-3p.
In the current study, we also found SZRD1 is the targeted gene of miR-128-3p. SZRD1, a highly conserved protein, is first found in cervical cancer, which functions as a tumor suppressor.35 Conversely, SZRD1 exerts the push roles on tumor development in the oligodendrogliomas, lymphoma, and glioma,21,36,37 which are in accordance with our results that overexpression of SZRD1 facilitated the tumorigenesis. Moreover, our study displayed that miR-128-3p mimic reduced the SZRD1 expression, whereas overexpression of SZRD1 reversed the function of miR-128-3p mimic on inhibition of glioma development, indicating that miR-128-3p was bound with SZRD1. This results were in line with a previous study, which revealed the interaction between miR-128-3p and SZRD1.21 The aforementioned findings manifested that miR-128-3p modulated the proliferation, and invasion of glioma cells via targeting SZRD1.
There were some limitations in the current study. First, the overall survival of mice among different groups was not examined in this study. In future, we will explore the effects of compounds on survival of mice including sh-FOXD3-AS1 and miR-128-3p. In addition, whether overexpression of FOXD3-AS1 influences on behaviors of glioma cell lines is unclear. Importantly, comparison of the growth rate and invasion among the glioma cell lines will be a potential research direction.
Conclusions
FOXD3-AS1 promoted proliferation, and invasion of glioma cells via regulation of the miR-128-3p/SZRD1 axis. Those findings clarified that the FOXD3-AS1/miR-128-3p/SZRD1 network may be an underlying mechanism for glioma development.
Ethics Approval and Informed Consent
The study was approved by the ethical committee of Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University (No. 2021-468), and all experiments conformed to Guide for the Care and Use of Laboratory Animals.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The study was funded by Major Science and Technology Innovation Project of Shandong Province (2020SFXGFY03-2).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Morgan LL. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2015;17(4):623–624. doi:10.1093/neuonc/nou358
2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1
3. Rich JN, Bigner DD. Development of novel targeted therapies in the treatment of malignant glioma. Nat Rev Drug Discov. 2004;3(5):430–446. doi:10.1038/nrd1380
4. Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee SU. Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev. 2017;18(1):3–9. doi:10.22034/APJCP.2017.18.1.3
5. Mohtashami E, Shafaei-Bajestani N, Mollazadeh H, et al. The current state of potential therapeutic modalities for glioblastoma multiforme: a clinical review. Curr Drug Metab. 2020;21(8):564–578. doi:10.2174/1389200221666200714101038
6. Pan W, Li G, Yang X, Miao J. Revealing the potential pathogenesis of glioma by utilizing a glioma associated protein-protein interaction network. Pathol Oncol Res. 2015;21(2):455–462. doi:10.1007/s12253-014-9848-9
7. Rajesh Y, Pal I, Banik P, et al. Insights into molecular therapy of glioma: current challenges and next generation blueprint. Acta Pharmacol Sin. 2017;38(5):591–613. doi:10.1038/aps.2016.167
8. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24(3):257–277. doi:10.1016/j.molmed.2018.01.001
9. Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):1015. doi:10.3390/cells8091015
10. Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. doi:10.1186/s12943-018-0812-2
11. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
12. Xin J, Zhao YH, Zhang XY, Tian LQ. LncRNA NFIA-AS2 promotes glioma progression through modulating the miR-655-3p/ZFX axis. Hum Cell. 2020;33(4):1273–1280. doi:10.1007/s13577-020-00408-9
13. Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2020;112(1):1021–1029. doi:10.1016/j.ygeno.2019.06.017
14. Zeng ZL, Zhu HK, He LF, et al. Highly expressed lncRNA FOXD3-AS1 promotes non-small cell lung cancer progression via regulating miR-127-3p/mediator complex subunit 28 axis. Eur Rev Med Pharmacol Sci. 2020;24(5):2525–2538. doi:10.26355/eurrev_202003_20520
15. Chen X, Gao J, Yu Y, Zhao Z, Pan Y. LncRNA FOXD3-AS1 promotes proliferation, invasion and migration of cutaneous malignant melanoma via regulating miR-325/MAP3K2. Biomed Pharmacother. 2019;120:109438. doi:10.1016/j.biopha.2019.109438
16. Wu Q, Shi M, Meng W, Wang Y, Hui P, Ma J. Long noncoding RNA FOXD3-AS1 promotes colon adenocarcinoma progression and functions as a competing endogenous RNA to regulate SIRT1 by sponging miR-135a-5p. J Cell Physiol. 2019;234(12):21889–21902. doi:10.1002/jcp.28752
17. Chen ZH, Hu HK, Zhang CR, et al. Down-regulation of long non-coding RNA FOXD3 antisense RNA 1 (FOXD3-AS1) inhibits cell proliferation, migration, and invasion in malignant glioma cells. Am J Transl Res. 2016;8(10):4106–4119.
18. Qu C, Yan C, Cao W, et al. miR-128-3p contributes to mitochondrial dysfunction and induces apoptosis in glioma cells via targeting pyruvate dehydrogenase kinase 1. IUBMB Life. 2020;72(3):465–475. doi:10.1002/iub.2212
19. Wang BC, Ma J. Role of microRNAs in malignant glioma. Chin Med J (Engl). 2015;128(9):1238–1244. doi:10.4103/0366-6999.156141
20. Yang X, Du H, Bian W, Li Q, Sun H. FOXD3‑AS1/miR‑128‑3p/LIMK1 axis regulates cervical cancer progression. Oncol Rep. 2021;45(5). doi:10.3892/or.2021.8013
21. Geng YB, Pan CC, Xu C, et al. Long non-coding RNA LINC00346 regulates proliferation and apoptosis by targeting miR-128-3p/SZRD1 axis in glioma. Eur Rev Med Pharmacol Sci. 2020;24(18):9581–9590. doi:10.26355/eurrev_202009_23046
22. Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–1157. doi:10.1007/s13311-018-0649-9
23. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
24. Zhao X, Li D, Huang D, et al. Risk-associated long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by repressing PARP1-mediated activation of CTCF. Mol Ther. 2018;26(3):755–773. doi:10.1016/j.ymthe.2017.12.017
25. Preker P, Almvig K, Christensen MS, et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39(16):7179–7193. doi:10.1093/nar/gkr370
26. Guan Y, Bhandari A, Xia E, Yang F, Xiang J, Wang O. lncRNA FOXD3-AS1 is associated with clinical progression and regulates cell migration and invasion in breast cancer. Cell Biochem Funct. 2019;37(4):239–244. doi:10.1002/cbf.3393
27. Ma WG, Shi SM, Chen L, Lou G, Feng XL. SP1-induced lncRNA FOXD3-AS1 contributes to tumorigenesis of cervical cancer by modulating the miR-296-5p/HMGA1 pathway. J Cell Biochem. 2021;122(2):235–248. doi:10.1002/jcb.29846
28. Wang X, Zhou J, Xu M, et al. A 15-lncRNA signature predicts survival and functions as a ceRNA in patients with colorectal cancer. Cancer Manag Res. 2018;10:5799–5806. doi:10.2147/CMAR.S178732
29. Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi:10.1016/j.bbrc.2005.07.030
30. Zhao L, Li R, Xu S, et al. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim Biophys Sin (Shanghai). 2018;50(2):171–180. doi:10.1093/abbs/gmx132
31. Pan J, Zhou C, Zhao X, et al. A two-miRNA signature (miR-33a-5p and miR-128-3p) in whole blood as potential biomarker for early diagnosis of lung cancer. Sci Rep. 2018;8(1):16699. doi:10.1038/s41598-018-35139-3
32. Liu T, Zhang X, Du L, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18(1):43. doi:10.1186/s12943-019-0981-7
33. Bendahou MA, Ibrahimi A, Boutarbouch M. Bioinformatics analysis of differentially expressed genes and miRNAs in low-grade gliomas. Cancer Inform. 2020;19:1176935120969692. doi:10.1177/1176935120969692
34. Kosti A, Barreiro R, Guardia GDA, et al. Synergism of proneurogenic miRNAs provides a more effective strategy to target glioma stem cells. Cancers (Basel). 2021;13(2):289. doi:10.3390/cancers13020289
35. Zhao N, Zhang G, He M, et al. SZRD1 is a novel protein that functions as a potential tumor suppressor in cervical cancer. J Cancer. 2017;8(11):2132–2141. doi:10.7150/jca.18806
36. Gladitz J, Klink B, Seifert M. Network-based analysis of oligodendrogliomas predicts novel cancer gene candidates within the region of the 1p/19q co-deletion. Acta Neuropathol Commun. 2018;6(1):49. doi:10.1186/s40478-018-0544-y
37. Carreras J, Hamoudi R, Nakamura N. Artificial intelligence analysis of gene expression data predicted the prognosis of patients with diffuse large B-cell lymphoma. Tokai J Exp Clin Med. 2020;45(1):37–48.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.