Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 16
LncRNA FOXD2-AS1 Increased Proliferation and Invasion of Lung Adenocarcinoma via Cell-Cycle Regulation
Authors Yuan Y , Yu P, Shen H, Xing G, Li W
Received 9 November 2022
Accepted for publication 6 January 2023
Published 3 February 2023 Volume 2023:16 Pages 99—109
DOI https://doi.org/10.2147/PGPM.S396866
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Yuan Yuan, Peng Yu, Huihua Shen, Guozhu Xing, Wu Li
Department of Cardiothoracic Surgery, Xinjiang Military Region General Hospital, Urumqi, People’s Republic of China
Correspondence: Yuan Yuan; Wu Li, Department of Cardiothoracic Surgery, Xinjiang Military Region General Hospital, No. 359, Youhao North Road, Urumqi, 830000, Xinjiang, People’s Republic of China, Tel +86-13899886276 ; +86-991-4992101, Email [email protected]; [email protected]
Background: Long non-coding RNA FOXD2 antisense RNA 1 (FOXD2-AS1) has been reported in many malignancies. However, the molecular mechanism of many actions is not clarified. This study was conducted to investigate the function of FOXD2-AS1 in lung adenocarcinoma and its molecular mechanism.
Methods: Bioinformatics and in vitro analysis including RT-qPCR, CFU, CCK8, Transwell, Cell Apoptosis and Cell Cycle Assay were used for the analysis of gene expression and related effects.
Results: It revealed increased expression of lncRNA FOXD2-AS1 in lung adenocarcinoma cell lines (A549 cells), and abundant expression of lncRNA FOXD2-AS1 was also observed in the acquired lung adenocarcinoma tissues. In vitro results showed that knockdown of lncRNA FOXD2-AS1 in A549 cells weakened cell proliferation, invasion and increased apoptosis. At the same time, we found that reducing the expression of lncRNA FOXD2-AS1 caused cell cycle arrest in the G1/S phase. Differential gene analysis of lung adenocarcinoma and adjacent normal tissues showed that the cell cycle and its related process regulation were significantly enriched. Gene Set Enrichment Analysis (GSEA) analysis showed that miR-206, miR-143, lL6-JAK-STAT3 signalling pathway, STAT3, E2F targets, EZH2, P53 signalling pathway and E2F3 targets interacting with lncRNA FOXD2-AS1 were also enriched.
Conclusion: This study demonstrates the role and mechanism of the lncRNA FOXD2-AS1 in lung adenocarcinoma and provides a better understanding for the treatment of lung adenocarcinoma, which indicates that interfering with lncRNA FOXD2-AS1 expression may be a novel strategy.
Keywords: lncRNA FOXD2-AS1, lung adenocarcinoma, non-small cell lung cancer, cell cycle, proliferation
Introduction
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer.1 In recent years, the development of surgery, chemotherapy, radiotherapy and targeted therapy has made certain progress in the treatment of NSCLC.2,3 However, early-stage NSCLC, especially lung adenocarcinoma with ground-glass nodules, is often difficult to detect because it has no obvious symptoms.2,4 Therefore, most lung adenocarcinoma patients are in the middle and late stages when they are diagnosed and have lost the best chance of surgery.5
LncRNA-FOXD2-AS1 was first reported in esophageal squamous cell carcinoma in 2015, and it was later confirmed to have carcinoid gene function in studies of liver cancer, gastric cancer, breast cancer and other diseases.6–9 FOXD2-AS1 promotes the occurrence and development of hepatocellular carcinoma by acting as a competitive endogenous RNA of miR-206. The expression level of Annexin A2 (ANXA2) in hepatocellular carcinoma is positively correlated with FOXD2-AS1, and miR-206 inhibits tumor development by downregulating the expression of ANXA2.10 However, the research on FOXD2-AS1 on lung cancer is less unknown, there is no widely recognized on the biological function and mechanism of FOXD2-AS1 in lung adenocarcinoma.
In this study, we intends to obtain the expression profiling data of lung adenocarcinoma and adjacent normal tissues from the GEO database. To further explore the relationship between LncRNA FOXD2-AS1 and key gene expression levels, and to further verify through in vitro experiments, to provide ideas and theoretical basis for in-depth research on the molecular mechanism of lung adenocarcinoma.
Materials and Methods
Data Acquisition and Processing
Gene expression profiling data numbered GSE168466 and GSE19804 were obtained from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) online database of the National Center for Biotechnology Information (NCBI). The data is based on the GPL23126 and GPL570 chip analysis platform, respectively. A total of 218 cases of data were collected, including 109 cases of non-small cell lung adenocarcinoma and 109 cases of corresponding paracancerous tissues. We then combine these data and re-analyze the data with R (version 4.2.0). The false positive rate was controlled by setting a P-value, and the false discovery rate (FDR) was calculated using the default Benjamini–Hochberg method.
Enrichment Analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO) and Reactome were used as bioinformatic tool that provides the comprehensive information on the gene function of individual genomic products based on the lung adenocarcinoma and corresponding paracancerous tissues. False discovery rate (FDR) was used to adjust the P-value. Genes with |log fold change (FC)| > = 1 and adjusted P < 0.05 were considered to be differentially expressed genes (DEGs). KEGG, GO and Reactome functional enrichment analyses were conducted using the R clusterProfiler package. Fisher’s test was used to identify the significant GO terms, KEGG pathways and Reactome terms, and P value <0.05 was considered statistically significant.
Gene Set Enrichment Analysis (GSEA)
GSEA is a promising and widely used software package, which derives gene sets to determine the different biological functions of the whole genes. The potential contribution of the whole altered genes in the lung adenocarcinoma was explored by the GSEA software (version 4.1.0). The GSEA software was downloaded from http://www.gsea-msigdb.org/gsea/index.jsp and was used to identify the potential function of the hub genes. FDR P <0.05 was used as the criterion for significant enrichment.
Clinical Sample Collection
The current study was approved by the Ethics Committee of The General Hospital of Xinjiang Military Region and conducted strictly in accordance with the Declaration of Helsinki. Signed informed consents were obtained from all participants prior to specimen collection, and the patient did not receive any treatment before collecting samples. A total of 6 patients with lung adenocarcinoma admitted at The General Hospital of Xinjiang Military Region were included in the current study. Initially, lung adenocarcinoma and adjacent tissue samples were collected from the subjects and immediately stored in a refrigerator at −80°C for subsequent experimentation.
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
The RNA in the 12 tissue samples was extracted with TRIzol reagent following the instruction of the manufacturer. The cDNA was synthesized using the Reverse Transcription System Kit (Applied Biosystems, USA) and 50 ng/uL of total RNA in a final volume of 20 uL were used and 260/280 ratio is between 1.9 and 2.1. And following reaction system was prepared: 25 μL PCR Master Mix, cDNA, ddH2O, Applied Biosystem 7500 Real-Time PCR system Perform the reaction on the top, optimize the primer and template concentration. All statistics were analyzed based on 2−ΔΔCt method. The primers were synthesized by Sangon (Shanghai, China) and the sequence is as follows: Actin-F, CTCCTGAGCGCAAGTACTCT; Actin-R, TACTCCTGCTTGCTGATCCAC; FOXD2-AS1-F, TGGACCTAGCTGCAGCTCCA; FOXD2-AS1-R, AGTTGAAGGTGCACACACTG.
Cell Culture and Transfection
The lung adenocarcinomic cell line A549 which is categorized as a lung adenocarcinoma was purchased from the American Type Culture Collection (ATCC) and cultured in DMEM containing 10% fetal bovine serum in a humidified incubator at 37°C with 5% CO2 in air. LncRNA FOXD2-AS1 overexpression plasmid (FOXD2-AS1), sh-FOXD2-AS1 and their negative controls were designed and synthesized by Sangon (Shanghai, China). Cells were transfected with indicated plasmids using Lipofectamine 2000, while cell density reached about 80%.
Colony-Forming Unit (CFU) Assay
The effects of overexpression LncRNA FOXD2-AS1 and sh-FOXD2-AS1 on the colony-forming ability of A549 cells were detected by CFU assay. Briefly, A549 cells were plated into six-well plates at a density of 1000 cells/well and incubated in a humidified CO2 incubator. After 12 days, wells were washed with PBS and fixed by using 4% paraformaldehyde (PFA). After crystal violet treatment, the dye was cleaned from the wells, and photos were taken with a luminometer device and colonies were counted. Colony formation rate = the colony formation number/the inoculated cell number × 100%. The experiment was implemented three repeated times.
CCK-8 Assay
Use the CCK-8 kit according to the manufacturer’s instructions. After incubate treated A549 cells for 24 h in 96-well plates. Add 10 μL of CCK8 solution to each well and then incubate for 2 h at 37 °C. Read the value at 450 nm with a microplate reader.
Transwell Assay
After cell transfection, 2 × 105 A549 cells were incubated for 24 h and resuspended in DMEM with 5% fetal bovine serum. Medium including 10% FBS was used to stimulate cell invasion. After 24 h, cells on the membrane surface were fixed with 4% paraformaldehyde for 15 min. 0.1% crystal violet solution was used to stain the invasion cells.
Cell Apoptosis Detection
Cell apoptotic ability was assayed by Annexin V-fluorescein isothiocyanate (FITC) cell apoptosis detection kit (Beyotime Biotechnology, Shanghai, China). The collected cells were suspended in 300 μL binding buffer and then incubated with 5 μL Annexin V-FITC solution at 4°C for 30 min. Afterward, 5 μL propidium iodide (PI) was used for further incubation for 5 min. Lastly, cell apoptosis was examined and analyzed with BD FACSCanto II equipped with BD FACSDiva software (Becton-Dickinson, USA).
Cell Cycle Analysis
According to the instructions of Cell Cycle Detection Kit, overexpression LncRNA FOXD2-AS1 and si-FOXD2-AS1 were collected into the centrifuge tube. After that, the cells were fixed with precooled 70% ethanol and stained with RNase A and propidium iodide (PI) for 30 min. Then, cell cycle distribution was analyzed using flow cytometry with CellQuest software.
Statistical Analysis
All experiments were performed in triplicate or more times, and all independent experiments were set for 3 times to take the average value. Data were presented as mean ± standard deviation, and statistical analysis was performed using GraphPad Prism 9.2.0 software. Student’s t-test or ANOVA followed by Tukey post-hoc test was used to compare differences. P < 0.05 was considered statistically significant.
Results
Identification of DEGs and the Expression of LncRNA FOXD2-AS1
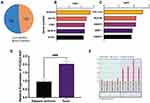
We performed a reanalysis of the GSE19804 dataset. With P<0.05, |logFC|>1 as the screening criteria, a total of 363 differentially expressed genes were obtained. Compared with lung adenocarcinoma and adjacent normal tissues, 173 genes were highly expressed and 190 genes were low expressed in lung adenocarcinoma (Figure 1A). Further, we found that the top 5 differentially up-regulated genes are SCGB1A1, ADH1B, CPB2, ZBTB16 and GSTA1, and the top 5 differentially down-regulated genes are COL11A1, MUC5B, HS6ST2, MMP1 and HS6ST2 (Figure 1B and C). These genes are all related to the regulation of cell proliferation and invasion.
To investigate LncRNA FOXD2-AS1 functions, we first detected the expression of LncRNA FOXD2-AS1 in lung adenocarcinomic cells by qRT-PCR. We found that LncRNA FOXD2-AS1 was up-regulated in lung adenocarcinoma (Figure 1D). As we known, there are many previous studies which detect the lncRNAs in lung adenocarcinoma by lncRNA microarray or others. According to microarray data (GSE168466) analysis, we found that LncRNA FOXD2-AS1 expression was increased in lung adenocarcinoma compared to adjacent normal tissues (Figure 1E). At the same time, LncRNA FOXD2-AS1 expression of NSCLC cases in TCGA database were analyzed. The results showed that LncRNA FOXD2-AS1 was highly expressed in 502 cases of Lung square cell cancer and 533 cases of Lung acinoma (Figure S1). The above results revealed that LncRNA FOXD2-AS1 was up-regulated in lung adenocarcinoma.
Downregulating LncRNA FOXD2-AS1 Inhibits the Proliferation and Invasion of in Lung Adenocarcinomic Cells
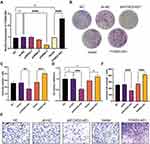
To characterize the function of LncRNA FOXD2-AS1, we firstly constructed three lentiviral vectors with LncRNA FOXD2-AS1 knocked down and overexpressed. Three stable transformants of A549 cell lines were obtained by down-regulation and overexpression of LncRNA FOXD2-AS1, which were identified by qPCR (Figure 2A). The proliferation of A549 cells with LncRNA FOXD2-AS1 and sh-FOXD2-AS1-3 was tested by Colony-forming unit (CFU) assay and CCK8 assay, and the CFU results showed that knocking down LncRNA FOXD2-AS1 could significantly inhibit colony-forming, however, overexpress of LncRNA FOXD2-AS1 promote the colony-forming of A549 cells (Figure 2B and C). CCK-8 results also showed that knocking down LncRNA FOXD2-AS1 could significantly inhibit the proliferation rate and overexpression of LncRNA FOXD2-AS1 enhanced the proliferation rate of A549 cells (Figure 2D). The results indicated that knocking down LncRNA FOXD2-AS1 significantly inhibited proliferation of lung adenocarcinomic cells. Transwell experiments showed that knocking down LncRNA FOXD2-AS1 significantly inhibited the invasion of A549 cells; however, overexpression of LncRNA FOXD2-AS1 enhanced the invasion of A549 cells (Figure 2E and F). In summary, downregulation of LncRNA FOXD2-AS1 inhibits the proliferation and invasion of lung adenocarcinomic cells.
LncRNA FOXD2-AS1 Affects the Cell Cycle Regulation in Lung Adenocarcinomic Cells
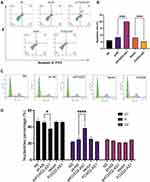
In order to find out the specific reason of the ability of LncRNA FOXD2-AS1 to inhibit cell proliferation and invasion, we examined the effect of LncRNA FOXD2-AS1 on apoptosis and cell cycle. The apoptosis rate of A549 cells with LncRNA FOXD2-AS1 and sh-FOXD2-AS1-3 was tested by flow cytometry, and the results showed that knocking down LncRNA FOXD2-AS1 could significantly enhance the apoptosis of A549 cells; however, overexpression of LncRNA FOXD2-AS1 inhibits the apoptosis of A549 cells (Figure 3A and B). We also observed that cell-cycle characterized by G1/S arrest in A549 cells after LncRNA FOXD2-AS1 was downregulating (Figures 3C and D). However, this effect was rescued by overexpression of LncRNA FOXD2-AS1 (Figures 3C and D). These results implied that LncRNA FOXD2-AS1 affects the cell cycle regulation in lung adenocarcinomic cells.
Enrichment Analysis Results
In order to further analyze the DEGs and to further explore the interaction of the DEGs and LncRNA FOXD2-AS1, we carried out GO, KEGG and Reactome enrichment for the DEGs. GO enrichment results showed that DEGs were mainly enriched in the biological process (Figure 4A). With regard to molecular function (MF), DEGs were primarily enriched in ATP-dependent microtubule motor activity (GO:1990939), microtubule motor activity (GO:0003777), microtubule binding (GO:0008017) and motor activity (GO:0003774) (Figure 4B). DEGs were also enriched in response to mitotic cell cycle process (GO:1903047), mitotic cell cycle (GO:0000278), cell cycle process (GO:0022402), cell cycle (GO:0007049), chromosome segregation (GO:0007059) and cell division (GO:0051301) in biological process (BP) (Figure 4C). It is also enriched in response to chromosome, centromeric region (GO:0000775), chromosomal region (GO:0098687) and kinetochore (GO:0000776) in Cellular Component (CC) (Figure 4D). KEGG analysis showed that DEGs were primarily enriched in cell cycle (ko04110), p53 signaling pathway (ko04115) and cellular senescence (ko04218) entries (Figure 4E and G). KEGG pathway analysis revealed that the DEGs were chiefly enriched in Cell cycle (ko04110) (Figure 4E and I). Moreover, Reactome pathway analysis revealed that the DEGs were chiefly enriched in Cell Cycle (R-HSA-1640170), Mitotic (R-HSA-69278) and Cell Cycle Checkpoints (R-HSA-69620) (Figure 4F and H). It can be seen from the above results that lung adenocarcinoma has an important relationship with the regulation of cell cycle.
Identification of the Potential Mechanism of LncRNA FOXD2-AS1 in Lung Adenocarcinoma Progression
To investigate the underlying mechanism of LncRNA FOXD2-AS1 in lung adenocarcinoma progression, we performed GSEA. GSEA is a computational method to explore whether a specific gene set is markedly enriched in a group of gene markers ranked by their relationship to a phenotype of interest. In the experiment, the expression profiles of samples were divided into lung adenocarcinoma tissue group and adjacent normal tissue group, and then analyzed based on hallmark gene sets, KEGG gene sets, GO gene sets, Reactome gene sets, miRNA target gene sets, transcription factor target gene sets and BioCarta gene sets. Several related gene sets, including miR206, miR143, lL6-JAK-STAT3 signaling, STAT3, E2F targets, EZH2, P53 signaling pathway and E2F3 targets were significantly enriched in the lung adenocarcinoma tissue group (Figure 5). It is important that all significantly enriched gene sets have a relationship with LncRNA FOXD2-AS1. Therefore, we can conclude that LncRNA FOXD2-AS1 is related to the proliferation and invasion of lung adenocarcinomic cells, and the regulation of cell cycle plays an important role in this process.
Discussion
Cell proliferation and invasion play a crucial role in maintaining normal tissue structure and function. More and more studies have shown that LncRNA affects gene transcription through molecular signals, thereby regulating biological functions such as cell proliferation and invasion.11 The disruption of the balance between abnormal cell proliferation and invasion leads to tumorigenesis. In recent years, more and more studies have shown that lncRNA plays a key regulatory role in a variety of tumors. Like protein targets, lncRNA can also serve as an oncogene.12
As a member of the lncRNA family, LncRNA FOXD2-AS1 was highly expressed in colorectal cancer, liver cancer, gastric cancer, breast cancer, gallbladder cancer and other malignant tumor tissues.9 In gastric cancer, abnormally high expression of FOXD2-AS1 is significantly correlated with tumor size and pathological stage, suggesting that patients have a poor prognosis. Yang et al found that high expression of FOXD2-AS1 can promote the proliferation of colorectal cancer cells. After interfering with the expression of FOXD2-AS1, the activity of intracellular EMT and Notch signaling pathways is reduced.13 In this study, through GEO database and lung adenocarcinoma samples, we found that LncRNA FOXD2-AS1 expression was increased in lung adenocarcinoma. At the same time, functional enrichment of DEGs has shown that cell cycle-related pathways including the process of mitotic cell cycle, chromosome segregation, cell division and nuclear division are all related to the process of lung adenocarcinoma. Through GSEA, we found that miR-206, miR-143, Stat3, EZH2, p53 and E2F3 were significantly enriched. However, some research has shown that LncRNA FOXD2-AS1 up regulate Annexin A2 (ANXA2) expression in part by “sponging” miR-206 and as an oncogene in hepatocellular carcinoma.10 LncRNA FOXD2-AS1 inhibits the expression of miR-143, then inhibits the resistance of human osteosarcoma cells to cisplatin, promotes their apoptosis and weakens their invasion and migration abilities.14 LncRNA FOXD2-AS1 also acted as a tumor inducer in gastric cancer through EphB3 inhibition by direct interaction with EZH2.15 It also could competitively sponge miR-760 and further upregulate the E2F3 expression to play a vital part in cholangiocarcinoma.16 We also noticed that ZBTB16 is highly expressed in lung adenocarcinoma (Figure 1). However, downregulation of ZBTB16 activated the phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) signaling pathway and restored the proliferation and invasiveness of gastric cancer cells.17 Combined with the highly expressed LncRNA FOXD2-AS1, we speculate that there is mutual regulation between LncRNA FOXD2-AS1 and ZBTB16, which affects the proliferation of lung adenocarcinoma.
Abnormal tumor cell cycle is affected by a variety of regulatory factors, including various LncRNAs. For example, in cervical cancer cells, LncRNA ANRIL affects cell proliferation by affecting the G2/M phase.18 Furthermore, Lnc ANRIL and/or UFC1 is an attractive target for drug development in tumor growth and aggressive proliferation of NSCLC.19 In lung cancer, silencing lncRNAs HOXA11-AS can cause cell cycle arrest in G0/G1 or G2/M phases.20 Cui et al found that the expression of LncRNA H19 in NSCLC down-regulated miR-107 and promoted cell cycle progression in NSCLC.21 We found that the number of cells in G1/S phase was significantly increased in A549 cells after knockdown of lncRNA FOXD2-AS1. Combined with CFU assay and proliferation experiments, the proliferation ability of A549 cells after knockdown of lncRNA FOXD2 was weakened, and the apoptosis was increased. This suggests that the mechanism by which lncRNA FOXD2-AS1 regulates the proliferation of A549 cells may be cell cycle G1/S arrest.
Conclusion
Taken together, the results of the present work revealed the role and potential mechanism of the lncRNA FOXD2-AS1 in lung adenocarcinoma and provided a better understanding for the treatment of lung adenocarcinoma, which indicated that interfering with lncRNA FOXD2-AS1 expression may be a novel strategy.
Acknowledgments
This work was supported by the Natural Science Foundation of Xinjiang Uygur Autonomous Region (2019D01A102).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Li C, Wang H, Jiang Y, et al. Advances in lung cancer screening and early detection. Cancer Biol Med. 2022;19(5):591–608. doi:10.20892/j.issn.2095-3941.2021.0690
3. Yang S, Huang Y, Zhao Q. Epigenetic alterations and inflammation as emerging use for the advancement of treatment in non-small cell lung cancer. Front Immunol. 2022;13:878740. doi:10.3389/fimmu.2022.878740
4. Nielsen AH, Fredberg U. Earlier diagnosis of lung cancer. Cancer Treat Res Commun. 2022;31:100561. doi:10.1016/j.ctarc.2022.100561
5. Ruparel M, Quaife SL, Dickson JL, et al. Lung screen uptake trial: results from a single lung cancer screening round. Thorax. 2020;75(10):908–912. doi:10.1136/thoraxjnl-2020-214703
6. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
7. Huang W, Li H, Yu Q, et al. LncRNA-mediated DNA methylation: an emerging mechanism in cancer and beyond. J Exp Clin Cancer Res. 2022;41(1):100. doi:10.1186/s13046-022-02319-z
8. Smolarz B, Zadrożna-Nowak A, Romanowicz H. The role of lncRNA in the development of tumors, including breast cancer. Int J Mol Sci. 2021;22(16):8427. doi:10.3390/ijms22168427
9. Hu Q, Tai S, Wang J. Oncogenicity of lncRNA FOXD2-AS1 and its molecular mechanisms in human cancers. Pathol Res Pract. 2019;215(5):843–848. doi:10.1016/j.prp.2019.01.033
10. Chang Y, Zhang J, Zhou C, et al. Long non-coding RNA FOXD2-AS1 plays an oncogenic role in hepatocellular carcinoma by targeting miR‑206. Oncol Rep. 2018;40(6):3625–3634.
11. Ghafouri-Fard S, Hussen BM, Gharebaghi A, Eghtedarian R, Taheri M. LncRNA signature in colorectal cancer. Pathol Res Pract. 2021;222:153432. doi:10.1016/j.prp.2021.153432
12. Liu Y, Zhang Y, Chen C, Li Y. lncRNA HIF1A-AS2: a potential oncogene in human cancers (Review). Biomed Rep. 2021;15(4):85. doi:10.3892/br.2021.1461
13. Yang X, Duan B, Zhou X. Long non-coding RNA FOXD2-AS1 functions as a tumor promoter in colorectal cancer by regulating EMT and notch signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(16):3586–3591.
14. Zhang QQ, Xu SL, Ding C, et al. LncRNA FOXD2-AS1 knockdown inhibits the resistance of human osteosarcoma cells to cisplatin by inhibiting miR-143 expression. Eur Rev Med Pharmacol Sci. 2021;25(2):678–686. doi:10.26355/eurrev_202101_24629
15. Xu TP, Wang WY, Ma P, et al. Upregulation of the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by epigenetically silencing EphB3 through EZH2 and LSD1, and predicts poor prognosis in gastric cancer. Oncogene. 2018;37(36):5020–5036. doi:10.1038/s41388-018-0308-y
16. Hu Z, Huang L, Wang W, et al. Long non-coding RNA FOXD2-AS1 promotes proliferation, migration, and invasion in cholangiocarcinoma through regulating miR-760/E2F3 axis. Dig Dis Sci. 2022;67(2):546–558. doi:10.1007/s10620-021-06876-9
17. Zhuang SH, Meng CC, Fu JJ, Huang J. Long non-coding RNA ELFN1-AS1-mediated ZBTB16 inhibition augments the progression of gastric cancer by activating the PI3K/AKT axis. Kaohsiung J Med Sci. 2022;38(7):621–632. doi:10.1002/kjm2.12548
18. Yao Y, Liang Y, Dong X, et al. Association of long non-coding RNAs (lncRNAs) ANRIL and MALAT1 polymorphism with cervical cancer. Pharmgenomics Pers Med. 2022;15:359–375. doi:10.2147/PGPM.S358453
19. Gupta S, Hashimoto RF. Dynamical analysis of a boolean network model of the oncogene role of lncRNA ANRIL and lncRNA UFC1 in non-small cell lung cancer. Biomolecules. 2022;12(3):420. doi:10.3390/biom12030420
20. Xia H, Niu Q, Ding Y, et al. Long noncoding HOXA11-AS knockdown suppresses the progression of non-small cell lung cancer by regulating miR-3619-5p/SALL4 axis. J Mol Histol. 2021;52(4):729–740. doi:10.1007/s10735-021-09981-1
21. Cui J, Mo J, Luo M, et al. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(10):12400–12409.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.