Back to Journals » Cancer Management and Research » Volume 12
LncRNA FEZF1-AS1 Sponges miR-34a to Upregulate Notch-1 in Glioblastoma
Authors Luo L, Zhang Y, He H, Chen C, Zhang B, Cai M
Received 29 November 2019
Accepted for publication 16 February 2020
Published 11 March 2020 Volume 2020:12 Pages 1827—1833
DOI https://doi.org/10.2147/CMAR.S240531
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Lun Luo,1 Yuan Zhang,2 Haiyong He,1 Chuan Chen,1 Baoyu Zhang,1 Meiqin Cai1
1Department of Neurosurgery, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong Province 510630, People’s Republic of China; 2Department of Obstetrics, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong Province 510630, People’s Republic of China
Correspondence: Lun Luo; Meiqin Cai
Department of Neurosurgery, The Third Affiliated Hospital of Sun Yat-sen University, No. 600 Tianhe Road, Tianhe District, Guangzhou, Guangdong Province 510630, People’s Republic of China
Tel +86-20-85253333
Email [email protected]; [email protected]
Introduction: LncRNA FEZF1-AS1 has been reported to be an oncogene in many types of cancer, while its role in glioblastoma (GBM) is unknown. This study aimed to investigate the potential involvement of FEZF1-AS1 in GBM.
Methods: FEZF1-AS1 expression in paired GBM and non-tumor tissues from GBM patients was determined by RT-qPCR. A 2-year follow-up was performed to analyze the prognostic value of FEZF1-AS1 for GBM. Cell transfections were performed to analyze the interactions between FEZF1-AS1, miR-34a and Notch-1. Transwell assay was performed to analyze the role of FEZF1-AS1, miR-34a and Notch-1 in regulating GBM cell invasion and migration.
Results: In this study, analysis of TCGA dataset revealed the upregulation of FEZF1-AS1 in GBM, and the overexpression of FEZF1-AS1 in GBM was further confirmed using GBM tissues from GBM patients included in this study. High levels of FEZF1-AS1 were correlated with poor survival. FEZF1-AS1 was predicted to form base pairing with miR-34a. However, overexpression of FEZF1-AS1 and miR-34a failed to affect the expression of each other. However, upregulation of Notch-1, a target of miR-34a, was observed after FEZF1-AS1 in GBM cells. Moreover, increased invasion and migration rates of GBM cells were observed after FEZF1-AS1 and Notch-1 overexpression. MiR-34a played an opposite role and reduced the effects of FEZF1-AS1 and Notch-1 overexpression.
Conclusion: FEZF1-AS1 may sponge miR-34a to upregulate Notch-1 in GBM, thereby promoting cancer cell invasion and migration.
Keywords: glioblastoma, FEZF1-AS1, miR-34a, Notch-1
Introduction
Glioblastoma (GBM) as a grade IV tumor is considered as the most aggressive brain tumors with high infiltration ability.1 GBM is a rare type of malignancy and only affect less than 5 out of 100,000 persons across populations.2 However, due to its extremely aggressive nature, most GBM patients will die within 2 years after initial diagnosis.3 GBM mainly affects the elderly. With the growing of aging population, it is estimated that the incidence of GBM will be increasing for a long term.4 To date, no cure is available for GBM. The unclear molecular pathogenesis continuously challenges the development of novel anti-GBM therapies.5 Therefore, in-depth investigations of the molecular pathways involved in GBM are always needed.
Previous oncological studies have identified a considerable number of molecular pathways involved in the pathogenesis.6,7 Some molecular players, such as microRNA miR-21 and PTEN signaling have been proven to be potential therapeutic targets for GBM.8,9 Notch homolog 1 (Notch-1) is single-pass transmembrane receptor that participates in cancer biology by regulating cancer cell behaviors.10 In GBM Notch-1 is overexpressed and promotes the invasion, migration and proliferation of cancer cells.11 In effect, certain tumor-suppressive miRNAs, such as miR-34a, can target Notch-1 to inhibit cancer development.12 Long non-coding RNA (lncRNA) FEZF1-AS1 has been characterized as an oncogenic lncRNA in several types of cancers, such as colorectal carcinoma and breast cancer.13,14 However, its involvement in GBM is unknown. We analyze TCGA dataset and observed the upregulation of FEZF1-AS1 in GBM. In addition, FEZF1-AS1 is predicted to interact with miR-34a. This study was therefore carried out to analyze the possible interaction between FEZF1-AS1 and miR-34a in GBM.
Materials and Methods
Patients and Follow-Up
This study was approved by Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University. This study enrolled a total of 60 GBM patients (38 males and 22 females, 55 to 74 years, 65.1 ± 4.5 years) from aforementioned hospital between January 2015 and December 2016. All patients were newly diagnosed cases. No recurrent GBM patients or previously diagnosed GBM patients were included. No anti-cancer therapies were initiated and patients complicated with other clinical disorders were excluded. Patients were treated with surgical resection, targeted therapy, radiotherapy, chemotherapy or their combinations. Biopsy was performed on all patients to collect both GBM tissues and adjacent (3 cm around tumors) non-tumor tissues from each patient. All patients signed written informed consent. Patients were followed up for 2 years to record their survival. All patients completed follow-up.
Cells and Cell Culture
U-251 and U-87 MG human GBM cell lines (ATCC) were used in this study. Cells were cultivated in a 5% CO2 incubator at 37ºC in a medium containing 10% FBS and DMEM medium. Cells were collected at 85% confluence to perform subsequent transfections.
Cell Transfections
FEZF1-AS1 and Notch-1 expression vectors were constructed using pcDNA3.1 vector. Negative control (NC) miRNA and miR-34a mimic were from Sigma-Aldrich. Lipofectamine 2000 (Invitrogen) was used to transfect 10 nM vector or 40 nM miRNA into 106 U-251 or U-87 MG cells following the protocol from Invitrogen. Controls (C) cells in all cases were untransfected cells. Cells were transfected with empty vector or NC miRNA to serve as NC group.
Dual-Luciferase Activity Assay
To explore the interaction between FEZF1-AS1 and miR-34a, pGL3 vector (Promega Corporation) was used to construct the vector of FEZF1-AS1. Lipofectamine 2000 (Invitrogen) was used to co-transfect 106 U-251 cells with FEZF1-AS1 vector + miR-34a mimic (miR-34a group) or FEZF1-AS1 vector + NC miRNA (NC group). Cells were cultivated under aforementioned conditions for 48 hrs, followed by the measurement of luciferase activity using Dual-Luciferase Assay System (BPS Bioscience)
RNA Preparations
RNA isolation from GBM and non-tumor tissues as well as U-251 cells was performed using Trizol reagent (Invitrogen). MiRNAs were harvested by precipitating RNA using 85% ethanol. All RNA samples were processed by gDNA eraser (Takara) to digest genomic DNA before use.
RT-qPCR Assays
BlazeTaq™ One-Step SYBR Green RT-qPCR Kit (Genecopoeia) was used to measure the expression levels of FEZF1-AS1 and Notch-1 with GAPDH as endogenous control. All steps were performed according to the manufacturer’s instructions. Measurement of the expression levels of mature miR-34a was performed using All-in-OneTM miRNA qRT-PCR Detection Kit (Genecopoeia) with U6 as endogenous control. All steps were completed following manufacturer’s instructions. Ct (ΔΔCt) method was used process Ct values of 3 replicates of reach experiment and to calculate relative gene expression levels.
Western Blot
Isolation of total protein from U-251 cells was performed using RIPA solution (Invitrogen). Protein concentration was measured by BCA method (Sigma-Aldrich). Following protein denaturation in boiling water for 10 min, proteins were separated in 10% SDS-PAGE gel. PVDF membrane was used for gel transfer, followed by blocking for 80 min at room temperature using 5% non-fat milk in PBS. Notch-1 (ab27526, Abcam) and GAPDH (ab9485, Abcam) rabbit primary antibodies were used to incubate with the membranes at 4°C for 12 hrs. After that, membranes were further incubated with HRP Goat Anti-Rabbit (IgG) secondary antibody (ab97051, Abcam) at 24°C for 2 hrs. ECL (Sigma-Aldrich) was used to process signals and Quantity One software was used to process all data.
Transwell Assays
Transwell filters (8 μm; BD Biosciences) were used to perform Transwell assays to analyze the effects of transfections on the invasion and migration of U-251 or U-87 cells. The upper Transwell chamber was filled with 3000 cells in 0.1 mL medium, and the lower chamber was filled with medium containing 20% FBS. It is worth noting that uncoated membranes were used for cell migration, while Matrigel-coated membranes were used for cell invasion. Cells were cultivated for 24 hrs under aforementioned methods. Lower surface of membranes was them stained with crystal violet (0.1%). Stained cells were observed under an optical microscope.
Statistical Analysis
All experiments were performed in at least three biological replicates. Data were expressed as mean values ± standard error. Paired t test was used to compare GBM and non-tumor tissues. Unpaired test was used to compare luciferase activity between 2 groups. Comparisons among multiple groups were performed by ANOVA (one-way) and Tukey’s test. The 60 patients were divided into high and low FEZF1-AS1 level groups (n=30) with its median expression level in GBM tissue as cutoff score. Survival curves were plotted based on follow-up data and were compared by Log-rank test. P<0.05 was statistically significant.
Results
FEZF1-AS1 Is Upregulated in GBM and Predicted Poor Survival
TCGA dataset was explored and it was observed that the expression level of FEZF1-AS1 was obviously higher in GBM tissues than in non-tumor tissues (0.33 vs 0.12). Expression levels of FEZF1-AS1 in GBM and non-tumor tissues from the 60 GBM patients were measured by performing qPCR. Compared to non-tumor tissues, expression levels of FEZF1-AS1 were significantly higher in GBM tissues (Figure 1A, p<0.001). Survival curves were plotted for both high and low FEZF1-AS1 level groups. Comparing to patients in low FEZF1-AS1 level group, overall survival rate of patients in high FEZF1-AS1 level group was significantly lower (Figure 1B).
FEZF1-AS1 and miR-34a Interacted with Each Other but Did Not Regulate Each Other’s Expression
IntaRNA 2.015 was used to predict the possible base pairs can be formed by FEZF1-AS1 and miR-34a. It was observed that FEZF1-AS1 and miR-34a can form strong base pairing (Figure 2A). To further analyze their interaction, dual-luciferase activity assay was performed by co-transfecting U-251 cells with FEZF1-AS1 vector + miR-34a mimic (miR-34a group) or FEZF1-AS1 vector + NC miRNA (NC group). Comparing to NC group, significantly lower relative luciferase activity was observed in miR-34a group (Figure 2B, p<0.05). U-251 cells were transfected with FEZF1-AS1 expression vector or miR-34a mimic, and the overexpression of FEZF1-AS1 and miR-34a was confirmed by RT-qPCR (Figure 2C). Compared to C and NC groups, overexpression of FEZF1-AS1 and miR-34a failed to affect the expression of each other (Figure 2D).
 |
Figure 2 FEZF1-AS1 and miR-34a interacted with each other but did not regulate each other’s expression. IntaRNA 2.015 was used to predict the possible base pairs can be formed by FEZF1-AS1 and miR-34a (A). Dual-luciferase activity assay was performed by co-transfecting U-251 cells with FEZF1-AS1 vector + miR-34a mimic (miR-34a group) or FEZF1-AS1 vector + NC miRNA (NC group). Luciferase activity was measured at 48 hrs post-transfection and compared (B). U-251 cells were transfected with FEZF1-AS1 expression vector or miR-34a mimic, and the overexpression of FEZF1-AS1 and miR-34a was confirmed by RT-qPCR (C). The effects of overexpression of FEZF1-AS1 and miR-34a on the expression of each other were also analyzed by RT-qPCR (D). Experiments were performed in 3 biological replicates and mean values were expressed, *p<0.05. |
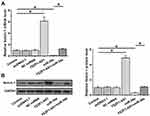
FEZF1-AS1 Upregulated Notch-1 Through miR-34a in U-251 Cells
FEZF1-AS1 and miR-34a interacted with each other but did not regulate each other’s expression. Therefore, it is reasonable to hypothesize that FEZF1-AS1 may serve as a sponge of miR-34a. To test this possibility, effects of FEZF1-AS1 and miR-34a on the expression of Notch-1, a target of miR-34a, were analyzed by qPCR (Figure 3A) and Western blot (Figure 3B). It observed that miR-34a overexpression led to the downregulated Notch-1 (p<0.05). In contrast, upregulation of Notch-1 was observed after FEZF1-AS1 (p<0.05). Moreover, FEZF1-AS1 overexpression led to the inhibited role of miR-34a (p<0.05).
FEZF1-AS1 Promoted GBM Cell Invasion and Migration Through miR-34a/Notch-1 Axis
Transwell assays were performed to analyze the effects of FEZF1-AS1, miR-34a and Notch-1 overexpression on the invasion (Figure 4A) and migration (Figure 4B) of U-251 cells. Increased invasion and migration rates of GBM cells were observed after FEZF1-AS1 and Notch-1 overexpression. MiR-34a played an opposite role and reduced the effects of FEZF1-AS1 and Notch-1 overexpression (p<0.05). To further confirm the role of FEZF1-AS1, miR-34a and Notch-1 in regulating GBM cell invasion and migration, transwell assay was also performed on another GBM cell line named U-87 MG. Similarly, FEZF1-AS1 and Notch-1 overexpression led to increased invasion (Supplementary Figure 1A, p<0.05) and migration (Supplementary Figure 1B, p<0.05) of U-87 MG cells. MiR-34a played an opposite role and reduced the effects of FEZF1-AS1 and Notch-1 overexpression (p<0.05).
Discussion
This study is the first to analyze the involvement of FEZF1-AS1 in GBM. We found that FEZF1-AS1 was upregulated in GBM and may interact with miR-34a/Notch-1 axis to participate in the regulation of cancer cell invasion and migration in GBM.
Previous studies have characterized FEZF1-AS1 as an oncogenic lncRNA in several types of cancers.13,14,16 For instance, FEZF1-AS1 is upregulated in breast cancer can target miR-30a/Nanog axis to increase cancer cell stemness.14 In another study, Zhao et al reported that FEZF1-AS1 was overexpressed in ovarian cancer can promote cancer cell proliferation and inhibited cancer cell apoptosis through the activation of JAK-STAT3 pathway.16 In this study, we observed the upregulation of FEZF1-AS1 in GBM. In addition, increased GBM cell invasion and migration rates of GBM cells were observed after FEZF1-AS1 overexpression. Therefore, FEZF1-AS1 also plays oncogenic roles in GBM.
It is estimated that the median survival time of GBM patients is only 11 months and only less than 5% of patients can survive longer than 2.5 years after initial diagnosis even after active treatment.3 Therefore, accurate prognosis of GBM may improve the survival by guiding the selection of therapies. In our study we showed that high levels of FEZF1-AS1 expression were closely correlated with the poor survival of GBM patients, indicating the potential application of FEZF1-AS1 as a prognostic marker for GBM. However, future studies with bigger sample size are needed to further test the reliability.
In a recent study, Kashat et al reported that miR-34a can target Notch-1 to inhibit the development of prostate cancer.12 Consistently, our study also observed the downregulation of Notch-1 in prostate cells after miR-34a overexpression. Therefore, miR-34a may also target Notch-1 in GBM. Our data suggested that FEZF1-AS1 might serve as an endogenous sponge of miR-34a to upregulate Notch-1, thereby promoting cancer cell invasion and migration. However, other mechanism may also exist and future studies are still needed.
Conclusion
In conclusion, FEZF1-AS1 is upregulated in GBM and may promote GBM cell invasion and migration through miR-34a/Notch-1 axis.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Alexander BM, Cloughesy TF. Adult glioblastoma. J Clin Oncol. 2017;35(21):2402–2409. doi:10.1200/JCO.2017.73.0119
2. Dobes M, Khurana VG, Shadbolt B, et al. Increasing incidence of glioblastoma multiforme and meningioma, and decreasing incidence of Schwannoma (2000–2008): findings of a multicenter Australian study. Surg Neurol Int. 2011;2:176. doi:10.4103/2152-7806.90696
3. Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107(2):359–364. doi:10.1007/s11060-011-0749-4
4. Babu R, Komisarow JM, Agarwal VJ, et al. Glioblastoma in the elderly: the effect of aggressive and modern therapies on survival. J Neurosurg. 2016;124(4):998–1007. doi:10.3171/2015.4.JNS142200
5. Bastien JI, McNeill KA, Fine HA. Molecular characterizations of glioblastoma, targeted therapy, and clinical results to date. Cancer. 2015;121(4):502–516. doi:10.1002/cncr.28968
6. Jhanwar-Uniyal M, Labagnara M, Friedman M, Kwasnicki A, Murali R. Glioblastoma: molecular pathways, stem cells and therapeutic targets. Cancers (Basel). 2015;7(2):538–555. doi:10.3390/cancers7020538
7. Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829–848. doi:10.1007/s00401-015-1432-1
8. Masoudi MS, Mehrabian E, Mirzaei H. MiR-21: a key player in glioblastoma pathogenesis. J Cell Biochem. 2018;119(2):1285–1290. doi:10.1002/jcb.v119.2
9. Banasavadi-Siddegowda YK, Russell L, Frair E, et al. PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene. 2017;36(2):263–274. doi:10.1038/onc.2016.199
10. Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer Biol Ther. 2002;1(5):466–476. doi:10.4161/cbt.1.5.159
11. Zhang X, Chen T, Zhang J, et al. Notch1 promotes glioma cell migration and invasion by stimulating beta-catenin and NF-kappaB signaling via AKT activation. Cancer Sci. 2012;103(2):181–190. doi:10.1111/j.1349-7006.2011.02154.x
12. Kashat M, Azzouz L, Sarkar SH, Kong D, Li Y, Sarkar FH. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am J Transl Res. 2012;4(4):432–442.
13. Chen N, Guo D, Xu Q, et al. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget. 2016;7(10):11271–11283. doi:10.18632/oncotarget.7168
14. Zhang Z, Sun L, Zhang Y, Lu G, Li Y, Wei Z. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J Cell Physiol. 2018;233(11):8630–8638. doi:10.1002/jcp.26611
15. Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45(W1):W435–W439. doi:10.1093/nar/gkx279
16. Zhao X, Cheng Z, Wang J. Long noncoding RNA FEZF1-AS1 promotes proliferation and inhibits apoptosis in ovarian cancer by activation of JAK-STAT3 pathway. Med Sci Monit. 2018;24:8088–8095. doi:10.12659/MSM.911194
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.