Back to Journals » Cancer Management and Research » Volume 13
LncRNA FENDRR Inhibits Gastric Cancer Cell Proliferation and Invasion via the miR-421/SIRT3/Notch-1 Axis
Authors Ma J, Zhao G, Du J, Li J, Lin G, Zhang J
Received 14 July 2021
Accepted for publication 9 November 2021
Published 14 December 2021 Volume 2021:13 Pages 9175—9187
DOI https://doi.org/10.2147/CMAR.S329419
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Jia Ma,1,* Gang Zhao,2,* Jia Du,1 Jiang Li,1 Guangshuai Lin,1 Jianfei Zhang1
1Department of Surgical Oncology, Shaanxi Provincial People’s Hospital, Xi’an, 710068, Shaanxi, People’s Republic of China; 2Department of Surgical Oncology, Pucheng County Hospital, Weinan, 715500, Shaanxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianfei Zhang
Department of Surgical Oncology, Shaanxi Provincial People’s Hospital, Xi’an, 710068, Shaanxi, People’s Republic of China
Tel +86-17391689521
Email [email protected]
Objective: This study aimed to investigate the regulatory effect of lncRNA fetal-lethal non-coding developmental regulatory RNA (FENDRR) on gastric cancer (GC) progression.
Methods: The expression levels of FENDRR in GC tissues and paracancerous tissues, as well as in gastric normal epithelial cell line and GC cell lines were detected. The Ad-FENDRR or si-FENDRR was transfected into AGS and SGC-7901 cells, and cell proliferation, invasion and apoptosis were determined. Online bioinformatics database predicted and screened miR-421 as a potential target of FENDRR, and SIRT3 was predicted as a target gene of miR-421. The pcDNA-SIRT3 or si-SIRT3 was transfected into AGS cells, and cell proliferation, invasion, apoptosis and Notch-1 protein expression were determined. Ad-FENDRR was transfected into AGS and SGC-7901 cells alone or together with miR-421 mimic to explore the effect of miR-421 on cells. The AGS cells transfected with Ad-FENDRR were injected into the armpits of nude mice to establish subcutaneous xenograft tumor model, and tumor growth was observed.
Results: FENDRR expression was downregulated in GC tissues and cell lines. Overexpression of FENDRR or SIRT3 inhibited tumor proliferation and invasion, and promoted apoptosis. The overexpression of Notch-1 reversed the inhibitory effect of SIRT3 on AGS cell. MiR-421 mimic reversed the inhibitory effect of FENDRR on the growth of AGS and SGC-7901 cells. Nude mice injected with FENDRR overexpressing AGS cells had smaller tumor volume and weight and weaker tumor cell proliferation ability.
Conclusion: FENDRR inhibits Notch-1 pathway to inhibit GC cell proliferation and invasion by upregulating SIRT3 expression via targeting miR-421.
Keywords: gastric cancer, lncRNA FENDRR, miR-421, SIRT3, Notch-1 pathway
Introduction
Gastric cancer accounts for more than 95% of gastric malignant tumors and is a common malignant tumor of the digestive tract worldwide. The incidence of gastric cancer is second only to the number one lung cancer, and the mortality rate is located after liver cancer and lung cancer.1 Despite the deepening of clinical diagnosis and treatment techniques and basic tumor research, the early detection and treatment rate of gastric cancer has increased, but the status of high mortality still exists. Therefore, it is urgent to study the molecular mechanism of the occurrence and development of gastric cancer, and to find more effective early diagnosis, prognostic markers and molecular therapeutic targets.
LncRNA is a class of nucleotide transcripts that lack protein-encoding functions and is approximately 200–100,000 nucleotides.2 LncRNA is involved in the regulation of gene expression at different levels, including transcriptional, post-transcriptional, translational, and post-translational.3 In the past decade, gastric cancer-related lncRNA studies have yielded significant results. LncRNA FENDRR was reported to be significantly downregulated in tumor tissues compared with normal tissues, and reduced FENDRR was associated with aggressive features of tumors.4 Xu et al showed that low FENDRR expression was associated with deeper tumor invasion, higher tumor stage, and lymph node metastasis, and mechanistic studies revealed that FENDRR overexpression inhibited gastric cancer cell invasion and migration by downregulating the expression of fibronectin 1 and matrix metalloproteinase 2/matrix metalloproteinase 9.5 Another study reported that downregulation of FENDRR expression reduced the drug resistance capacity of multi drug resistant gastric cancer cells, whereas upregulation of FENDRR expression had the opposite effect, and this was mediated through the miR-4700-3p and FOXC2 axis.6
MicroRNAs (miRNAs) are small non-coding nucleotides and are specifically expressed in various types of cells and tissues. As important post-transcriptional regulatory molecules, microRNAs can regulate the expression of target genes by binding to multiple target mRNAs. Dysregulation of miRNAs has been identified as a crucial factor in carcinogenesis. There is a study suggesting that miR-421, which is highly expressed in the plasma of gastric cancer patients, may serve as a potential diagnostic biomarker for precancerous lesions and early gastric cancer.7 Ge et al indicated that the overexpression of miR-421 promoted gastric cancer cell invasion, inhibited apoptosis, and induced cisplatin resistance, and E-cadherin and caspase-3 were identified as targets of miR-421.8 Claudin 11 was identified as a direct target of miR-421, and downregulation of miR-421 expression inhibited gastric cancer cell proliferation and metastasis and induced G1/S phase cell cycle arrest, and similar results were observed after Claudin 11 protein overexpression.9
SIRT3 is a key NAD+-dependent protein deacetylase in the mitochondria of mammalian cells, functioning to prevent cell aging and transformation via regulation of mitochondrial metabolic homeostasis. At present, studies have shown that SIRT3 plays an important role in gastric carcinogenesis and progression. Huang et al found that gastric cancer patients with more SIRT3 expression had a better prognosis, suggesting that SIRT3 could be an independent prognostic marker for overall survival and a tumor suppressor in the progression of gastric cancer.10 Another study showed that overexpression of SIRT3 inhibited the proliferation ability and colony formation number of gastric cancer cells, inhibited Notch-1 expression, and interference with SIRT3 expression abolished these changes, suggesting that SIRT3 inhibits gastric cancer cell growth by downregulating Notch-1.11
In this study, we explore the specific mechanism of FENDRR in the occurrence and development of gastric cancer by investigating its effect on gastric cancer cell proliferation and migration, aiming to provide new reference ideas for the diagnosis and treatment of gastric cancer.
Materials and Methods
Cell Culture and Reagents
The human GC cell lines AGS, MKN-45 and SGC-7901 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in RPMI 1640 medium (Gibco, Rockville, MD) containing 10% fetal bovine serum and 100 U/mL penicillin and 100 μg/mL streptomycin (Sigma, St. Louis, MO, USA) with 5% CO2 at 37°C. MiR-421 mimic, negative control oligonucleotides (NC mimic), FENDRR adenovirus vector (Ad-FENDRR) and empty vector (vector) were purchased from RiboBio Co., Ltd (Guangzhou, China) and transfected using RiboBio Transfection Kit (RiboBio Co., Ltd). Small interfering RNA against FENDRR (siFENDRR) and scramble siRNA against FENDRR (NC siRNA) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All transfection reagent transfected into cells using Lipofectamine 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Xenograft Experiments
BALB/c nude mice (aged 4 weeks, weight 20–22 g, Beijing Vital River laboratory animal technology company, China) were housed in a specific pathogen-free environment under the condition of 12-h light/12-h dark cycle, free access to food and water, and acclimatized to their surroundings for three days. These nude mice were randomly divided into three groups (n = 8 per group) including control group (AGS cells), vector group (A549 cells transfected with vector) and Ad-FENDRR group (A549 cells transfected with Ad-FENDRR). AGS cells (2 × 106) transfected with vector or Ad-FENDRR were injected subcutaneously into the armpits of nude mice. The tumor volume was closely observed and measured on weeks 1, 2, 3 and 4 after injection. On weeks 4 (28 d), nude mice were euthanized, and tumor tissues were collected and weighed. All experiments were conducted in accordance with the Animal Ethics Committee of Shaanxi Provincial People’s Hospital.
RT-qPCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as the instruction for manufacture. Reverse transcription was performed by using a PrimeScript RT reagent kit (Takara Biotechnology, Dalian, China). Real-time PCR analysis was conducted with the SYBR Premix Ex Taq II (Takara, Dalian, China) on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) under the following conditions: 95°C for 1 min, and then 95°C for 20 s, 56°C for 10 s and 72°C for 15 s for 35 cycles. The primers used in this study were synthesized from Sangon Biotech (Shanghai, China). The relative expression levels were normalized by using the 2−ΔΔCt method.
CCK-8 Assay
The Cell Counting Kit-8 (Beyotime Biotechnology, Shanghai, China) was used to assess the cell proliferation. Cells were grown to sub-confluence and harvested into Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). The cells were then seeded at a concentration of 1 × 104 cells/well on commercial 96-well plates and allowed to adhere overnight at 37°C. 10 μL of thawed CCK-8 solution was added to each well. After the plates were incubated for 2 h at the same incubator conditions, the absorbance at 450 nm was detected with a micro-plate analyzer (Molecular Devices, Sunnyvale, CA, USA).
Transwell Invasion Assay
Transwell invasion assay was performed by using Transwell chamber (8 µm pore size; Millipore Corporation, USA) coated with Matrigel (BD Biosciences, USA). A total of 1 × 105 AGS or SGC-7901 cells with serum-free culture medium were seeded into the upper chamber, while culture medium containing 20% FBS was added to the lower chamber. After incubation for 24 h at 37°C, the cells that stayed in the top chamber were carefully removed by using a cotton swab, and the cells in the lower chamber were fixed with 70% ethanol for 10 min and stained with 0.1% crystal violet for 15 min. The number of invasive cells was counted under a microscope light microscope (Olympus, Tokyo, Japan).
Luciferase Reporter Gene Assay
StarBase (http://starbase.sysu.edu.cn/) was used to predict the potential target genes of FENDRR, and miR-421 is screened as a research object. The wild type and mutant sequences of KCNQ1OT1 were cloned into the PGL3 luciferase reporter vector (Promega, USA), respectively. Cells were seeded in 24-well plates, when growing to approximately 70% confluence, and co-transfected with luciferase plasmids and miR-421 mimic or NC-mimic by using Lipofectamine 2000. After 48 h transfection, the luciferase activity was determined by Multiskan FC Microplate Reader (Thermo Fisher Scientific, Waltham, MA, USA).
Flow Cytometry
Cell apoptosis assay was performed by using an Annexin V-FITC-PI Apoptosis Detection kit (BD Biosciences, San Jose, CA) according to the manufacturer’s instructions. Briefly, cells (2 × 105) were resuspended in 200 μL of binding buffer. Then, 10 μL of Annexin V-FITC and 10 μL of PI were added, and the cells were incubated in the dark at 4°C for 15 min. Cell apoptosis was detected using a Flow Cytometry (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer’s instruction.
Western Blotting
Proteins were extracted from GC cells and tumor tissues by using RIPA lysis buffer (Beyotime, Shanghai, China). Total protein concentration was measured by BCA Protein Assay Kit (Beyotime, Shanghai, China). The proteins were then separated in SDS-PAGE under the following conditions: 70 V for 30 min, followed by 120 V for 90 min. And then the protein bands were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA) at 300 mA for 2 h. After being blocked with 5% nonfat milk for 2 h at room temperature, the membranes were incubated overnight at 4°C with the following primary antibodies: GAPDH antibody (1:2500, Abcam, ab9485), SIRT3 antibody (1:1000, Abcam, ab217319), Notch-1 antibody (1:1500, Abcam, ab52627). Then, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:2000, Abcam, ab6721) for 1 h. The protein bands were visualized with ECL detection reagents and analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Statistical Analysis
All statistical analyses were performed using the SPSS software (ver. 21.0; SPSS, Chicago, IL). The quantitative data derived from three independent experiments are expressed as mean ± SEM. Significance was determined by one-way ANOVA or t-test. Values of P<0.05 were considered statistically significant.
Results
FENDRR Was Downregulated in GC Tissues and Cell Lines
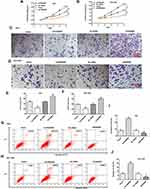
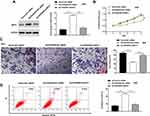
In this research, we evaluated the expression of FENDRR in 35 paired GC tissues and adjacent tissues and found that the FENDRR was markedly downregulated in GC tissues (Figure 1A). Meanwhile, RT-qPCR results showed that compared with the normal gastric epithelial cell line GES, FENDRR was significantly downregulated in GC cell lines (Figure 1B). Furthermore, transfection of a FENDRR overexpression vector (Ad-FENDRR) in GC cells promoted FENDRR expression (Figure 1C), and transfection of small interfering RNA against FENDRR inhibited FENDRR expression (Figure 1D). Analyzing the association between FENDRR expression levels and overall survival, we found that patients with high FENDRR expression had a better overall survival than those with low FENDRR expression (Figure 1E). The above results indicated that FENDRR expression was upregulated in GC tissues and cell lines, and its upregulation may be associated with GC progression.
FENDRR Inhibited GC Cell Proliferation and Invasion and Promoted Apoptosis
To further explore the biological effect of FENDRR on gastric cancer development, we bidirectionally regulated FENDRR expression in AGS and SGC-7901 cells. CCK-8 assay results showed that overexpression of FENDRR inhibited AGS and SGC-7901 cell proliferation, and transfection of siFENDRR enhanced AGS (Figure 2A) and SGC-7901 cell proliferation (Figure 2B). Moreover, the overexpression of FENDRR inhibited AGS and SGC-7901 cell invasion, and the transfection of siFENDRR promoted AGS (Figure 2C and E) and SGC-7901 cell invasion (Figure 2D and F). Flow cytometry results indicated that overexpression of FENDRR promoted apoptosis in AGS and SGC-7901 cells, and transfection of siFENDRR inhibited apoptosis in AGS (Figure 2G and I) and SGC-7901 cells (Figure 2H and J). The above results indicated that overexpression of FENDRR inhibited GC cell proliferation and invasion and promoted apoptosis, and knockdown of FENDRR promoted GC cell proliferation and invasion and inhibited apoptosis.
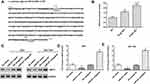
MiR-421 Targeted FENDRR and SIRT3
We predicted the potential binding region between FENDRR and miR-421 through the online bioinformatics database (http://starbase.sysu.edu.cn/). The wild and mutant sequences of FENDRR are shown in Figure 3A. Next, luciferase reporter gene analysis verified the targeting relationship between miR-421 and FENDRR (Figure 3B). RNA pull-down assay results showed that miR-421 probe was significantly enriched in FENDRR (Figure 3C). Furthermore, transfection of FENDRR inhibited miR-421 expression in AGS and SGC-7901 cells, and knockdown of FENDRR promoted miR-421 expression (Figure 3D).
In addition, we found that miR-421 directly targeted the 3ʹUTR of SIRT3. Since miR-421 has three binding sites with STAT3, we compared the mRNA sequence of SIRT3 and performed double mutations (Figure 4A). Luciferase reporter gene analysis revealed that wild-type SIRT3 significantly reduced luciferase activity (Figure 4B). Western blotting showed that miR-421 mimic inhibited SIRT3 expression in AGS and SGC-7901 cells, and miR-421 antagomir promoted SIRT3 expression in AGS (Figure 4C and D) and SGC-7901 cells (Figure 4C and E). In addition to SIRT3, we also examined the expression levels of several other downstream targets of miR-421, including KLF6, TIMP2 and BTG2, in tumor tissues and paracancerous tissues, as well as in gastric cancer cell lines and normal gastric epithelial cells, and showed that the above four genes were downregulated in both tumor tissues and human gastric cancer cell lines, but SIRT3 was the most significantly downregulated (Supplementary Figure 1). The above results indicate that miR-421 is a target gene of FENDRR and that miR-421 targets the 3ʹUTR of SIRT3.
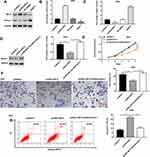
SIRT3 Inhibited GC Cell Proliferation and Invasion by Inhibiting Notch-1 Pathway
The pcDNA-SIRT3 and si-SIRT3 were transfected into AGS cells, respectively. Western blotting showed that transfected pcDNA-SIRT3 upregulated SIRT3 expression (Figure 5A and B) and inhibited Notch-1 expression (Figure 5A and C). Next, AGS cells were transfected with pcDNA-SIRT3 alone or together with pcDNA-Notch-1. We found that transfection of pcDNA-SIRT3 inhibited Notch-1 protein expression, and transfection of pcDNA-Notch-1 reversed this effect (Figure 5D). Next, CCK-8 assay results showed that pcDNA-Notch-1 reversed the inhibitory effect of SIRT3 on cell proliferation (Figure 5E). Transwell invasion assay results indicated that pcDNA-Notch-1 reversed the inhibitory effect of SIRT3 on cell invasion (Figure 5E). Flow cytometry results showed that pcDNA-Notch-1 reversed the promotion effect of SIRT3 on cell apoptosis (Figure 5G). The above results indicated that overexpression of SIRT3 inhibited GC cell proliferation and invasion and promoted apoptosis, and transfection of pcDNA-Notch-1 reversed the inhibitory effect of pcDNA-SIRT3 on GC cell growth.
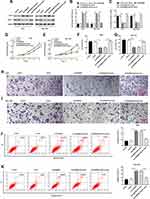
FENDRR Inhibited GC Cell Proliferation and Invasion by Targeting miR-421
To further explore the regulatory role of miR-421, the Ad-FENDRR was transfected into AGS and SGC-7901 cells alone or together with miR-421 mimic. Western blotting results showed that miR-421 mimic reversed the promoting effect of FENDRR on SIRT3 expression (Figure 6A and B) as well as the inhibitory effect on Notch-1 expression (Figure 6A and C). The results of CCK-8 assay showed that miR-421 mimic reversed the inhibitory effect of FENDRR on cell proliferation (Figure 6D and E). Furthermore, FENDRR inhibited cell invasion, whereas miR-421 mimic reversed the inhibitory effect of FENDRR on cell invasion (Figure 6F–I). Flow cytometry analysis showed that AGS (Figure 6J) and SGC-7901 (Figure 6K) cell apoptosis increased after transfection with Ad-FENDRR, while significantly decreased after co-transfection with miR-421 mimic and Ad-FENDRR. The above results indicated that overexpression of FENDRR inhibited GC cell proliferation and invasion and promoted apoptosis, and transfection of miR-421 mimic reversed the inhibitory effect of Ad-FENDRR on GC cell growth, suggesting that FENDRR may regulate GC cell growth through miR-421.
FENDRR Inhibited GC Cell Proliferation and Invasion by Upregulating SIRT3 Expression
The Ad-FENDRR was transfected into AGS cells alone or together with siSIRT3, and the results indicated that overexpression of FENDRR promoted SIRT3 protein expression (Figure 7A), inhibited cell proliferation (Figure 7B) and invasion (Figure 7C), and promoted cell apoptosis (Figure 7D), and transfection of siSIRT3 reversed the inhibitory effect of Ad-FENDRR on tumor cell growth. The above results indicated that FENDRR inhibited GC cell proliferation and invasion by upregulating SIRT3 expression.
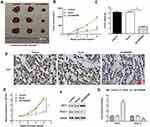
Overexpression of FENDRR Inhibited Tumor Progression in vivo
AGS cells transfected with vector or overexpressing FENDRR were injected subcutaneously into the armpits of nude mice to construct a subcutaneous xenograft tumor model to explore the ability of FENDRR to induce tumor formation in vivo. The representative tumor images are shown in Figure 8A. Furthermore, the tumors formed in nude mice injected with FENDRR overexpressing AGS cells were smaller in volume (Figure 8B) and weight (Figure 8C). Besides, we detected the expression of Ki67, a marker of cell proliferation, in tumor tissues by immunohistochemistry and found that the number of Ki67 positive cells was reduced in tumors formed induced by FENDRR overexpressing AGS cells compared to those formed by injection of AGS cells (Figure 8D). RT-qPCR results showed that overexpression of FENDRR inhibited miR-421 expression in tumor tissues (Figure 8E). Western blotting showed that SIRT3 protein expression was increased and Notch-1 protein expression was decreased in tumor tissues after overexpression of FENDRR (Figure 8F and G). The above results indicated that overexpression of FENDRR was able to inhibit tumor growth in nude mice, which was mediated by increasing SIRT3 expression via targeted inhibition of miR-421.
Discussion
Gastric cancer is one of the major public health problems worldwide. Despite the great improvement in the clinical treatments, including chemotherapy, radiotherapy and targeted therapies, GC is still the main cause of cancer-related deaths globally, and its overall prognosis is still poor. Recently, new evidence has shown that lncRNA and miRNA are involved in tumorigenesis and metastasis. For example, FENDRR expression is down-regulated in breast cancer tissues, and low levels of FENDRR are associated with shorter overall survival in breast cancer patients.12 In addition, FENDRR low expression was associated with a poor overall survival in patients with NSCLC and related to advanced stages and lymphatic metastasis.13
MiR-421 is a currently discovered miRNA associated with tumor progression, its expression is up-regulated in various tumor tissues, such as gastric cancer, glioma and osteosarcoma, and it can regulate the proliferation and migration process of tumor cells. Studies have shown that miR-421 is highly expressed in liver cancer and may be involved in the progression and metastasis of liver cancer.14 In this study, the expression level of miR-421 in gastric cancer cell lines was higher than that of normal gastric epithelial cells, and down-regulation of miR-421 could inhibit the proliferation of gastric cancer cells, suggesting that miR-421 may play a promoting role in the malignant progression of gastric cancer.
The biological role of miRNA is related to the expression of target proteins. This study shows that SIRT3 is negatively regulated by miR-421, and the action mechanism of miR-421 may be related to SIRT3. SIRT3 belongs to class III histone deacetylase, which can stimulate ribosyltransferase activity and deacetylase activity after being stimulated by relevant signals, affecting the expression of the target protein. Previous studies have shown that SIRT3 may play a bidirectional role for oncogenes and tumor suppressor gene.15 Li et al believed that abnormal expression of SIRT3 could reverse p53-mediated cell cycle arrest and promote tumor cell proliferation.16 In a functional study of SIRT3 as a tumor suppressor gene, Kim et al reported that SIRT3 gene knockout mice were prone to ER-positive breast cancer.17 Xiao et al observed down-regulated SIRT3 expression in lung adenocarcinoma cells, and overexpression of SIRT3 could induce apoptosis.18
The Notch gene was first discovered in Drosophila in 1917; then, it has shown that the Notch signaling pathway is an evolutionarily conserved signaling pathway, that is ubiquitous in mammalian cells and maintains cell proliferation.19 The activation of abnormal Notch signaling pathway is associated with the development of tumors. Among them, Notch-1 is the most studied receptor. Han et al showed that the cell invasion and migration ability was significantly inhibited by silencing the expression of Notch-1 gene by siRNA in HepG2 and SMMC7721.20 Feng et al reported that Notch-1 was highly expressed in gastric cancer, and the expression level of Notch-1 was correlated with tumor invasion depth, lymph node metastasis and long-term survival, suggesting that tumor progression and poor prognosis.21 These reported studies have further supported our experimental results.
In conclusion, FENDRR may be an inhibitor of gastric cancer progression and metastasis, and targeting FENDRR may be a pathway for the treatment of gastric cancer. Up-regulation of FENDRR may affect the proliferation, migration, invasion and apoptosis of gastric cancer cells by inhibiting miR-421 targeted regulation of SIRT3 expression and activation of the Notch-1 pathway, which provides a reference for studying the regulation mechanism of FENDRR in gastric cancer.
Abbreviations
lncRNAs, long non-coding RNAs; GC, gastric cancer; FENDRR, fetal-lethal non-coding developmental regulatory RNA; SIRT3, sirtuin 3; RUNX1, runt-related transcription factor 1; CALN1, calcium nutrient protein 1; NF-κB, nuclear transcription factor-κB; DMEM, Dulbecco’s `modified Eagle’s medium; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; PVDF, polyvinylidene difluoride; HRP, horseradish peroxidase.
Data Sharing Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.
Acknowledgments
We acknowledge and appreciate our colleagues for their valuable efforts and comments on this paper.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the key R & D program of Shaanxi Province (2020SF-221).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Cao Q, Liu F, Ji K, et al. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017;36(1):29. doi:10.1186/s13046-017-0499-z
2. Hao NB, He Y-F, Li X-Q, Wang K, Wang R-L. The role of miRNA and lncRNA in gastric cancer. Oncotarget. 2017;8(46):81572–81582. doi:10.18632/oncotarget.19197
3. Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. 2018;233(5):4044–4055.
4. Luo T, Zhao J, Lu Z, et al. Characterization of long non-coding RNAs and MEF2C-AS1 identified as a novel biomarker in diffuse gastric cancer. Transl Oncol. 2018;11(5):1080–1089. doi:10.1016/j.tranon.2018.06.007
5. Xu TP, Huang MD, Xia R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi:10.1186/s13045-014-0063-7
6. Liu H, Zhang Z, Han Y, et al. The FENDRR/FOXC2 axis contributes to multidrug resistance in gastric cancer and correlates with poor prognosis. Front Oncol. 2021;11:634579. doi:10.3389/fonc.2021.634579
7. Chen J, Wu L, Sun Y, et al. Mir-421 in plasma as a potential diagnostic biomarker for precancerous gastric lesions and early gastric cancer. PeerJ. 2019;7:e7002. doi:10.7717/peerj.7002
8. Ge X, Liu X, Lin F, et al. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget. 2016;7(17):24466–24482. doi:10.18632/oncotarget.8228
9. Yang P, Zhang M, Liu X, Pu H. MicroRNA-421 promotes the proliferation and metastasis of gastric cancer cells by targeting claudin-11. Exp Ther Med. 2017;14(3):2625–2632. doi:10.3892/etm.2017.4798
10. Huang KH, Hsu CC, Fang WL, et al. SIRT3 expression as a biomarker for better prognosis in gastric cancer. World J Surg. 2014;38(4):910–917. doi:10.1007/s00268-013-2359-0
11. Wang L, Wang WY, Cao LP. SIRT3 inhibits cell proliferation in human gastric cancer through down-regulation of Notch-1. Int J Clin Exp Med. 2015;8(4):5263–5271.
12. Yan L, Wenwen Z, Pengying L, et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Onco Targets Ther. 2018;11:1403–1412.
13. Gong F, Dong D, Zhang T, Xu W. Long non-coding RNA FENDRR attenuates the stemness of non-small cell lung cancer cells via decreasing multidrug resistance gene 1 (MDR1) expression through competitively binding with RNA binding protein HuR. Eur J Pharmacol. 2019;853:345–352.
14. Zhao YY, Mao MW, Zhang WJ, et al. Expanding RNA binding specificity and affinity of engineered PUF domains. Nucleic Acids Res. 2018;46(9):4771–4782. doi:10.1093/nar/gky134
15. Wang JX, Yi Y, Li YW, et al. Down-regulation of sirtuin 3 is associated with poor prognosis in hepatocellular carcinoma after resection. BMC Cancer. 2014;14(1):297. doi:10.1186/1471-2407-14-297
16. Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PloS one. 2010;5(5):e10486.
17. Kim HS, Jacobs PK. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17(1):41–52. doi:10.1016/j.ccr.2009.11.023
18. Xiao K, Jiang J, Wang W. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncol Rep. 2013;30(3):1323–1328. doi:10.3892/or.2013.2604
19. Hong K-J, Wu D-C, Cheng K-H, Chen L-T, Hung W-C. RECK inhibits stemness gene expression and tumorigenicity of gastric cancer cells by suppressing ADAM-mediated Notch1 activation. J Cell Physiol. 2014;229(2):191–201. doi:10.1002/jcp.24434
20. Han B, Liu SH, Guo WD, et al. Notch1 downregulation combined with interleukin-24 inhibits invasion and migration of hepatocellular carcinoma cells. World journal of gastroenterology. 2015;21(33):9727–9735.
21. Feng Y-Z, Shiozawa T, Miyamoto T, et al. Overexpression of Hedgehog signaling molecules and its involvement in the proliferation of endometrial carcinoma cells. Clin Cancer Res. 2007;13(5):1389–1398. doi:10.1158/1078-0432.CCR-06-1407
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.