Back to Journals » OncoTargets and Therapy » Volume 12
LncRNA FENDRR attenuates colon cancer progression by repression of SOX4 protein
Received 24 November 2018
Accepted for publication 17 April 2019
Published 30 May 2019 Volume 2019:12 Pages 4287—4295
DOI https://doi.org/10.2147/OTT.S195853
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Jianchao Liu, Wenfeng Du
Department of Gastroenterological Surgery, Liaocheng People’s Hospital, Liaocheng Clinical School of Taishan Medical University, Liaocheng, Shandong Province, 252000, People’s Republic of China
Background and purpose: Homo sapiens FOXF1 adjacent noncoding developmental regulatory RNA (FENDRR) is a novel long noncoding RNA (lncRNA) exerting important effects on transcriptional and post-transcriptional regulation. The purpose of this study was to investigate the potential role of FENDRR in colon cancer.
Methods: Multiple cellular and molecular biology experiments were performed in the present study, such as CCK-8, Western blot, immunohistochemistry, confocal immunofluorescent and animal studies.
Results: We determined that attenuation of FENDRR was a frequent event in colon cancer tissues and colon cancer cell lines, in contrast to their normal counterparts. Low levels of FENDRR were associated with the clinical stages and poor prognosis. Moreover, ectopic expression of FENDRR repressed colon cancer cell viability, invasion and epithelial–mesenchymal transition. Furthermore, through a series of in vitro and in vivo assays, we reported the discovery of FENDRR modulating the expression of SOX4 protein, and hence in the progression of colon cancer.
Conclusion: Based on these data, we demonstrated that FENDRR may function as a tumor-suppressor gene by repressing SOX4 and as a potential therapeutic target for colon cancer.
Keywords: FENDRR, SOX4, colon cancer, progression
Introduction
As one of the most prevalent malignancies worldwide, colon cancer accounts for one-tenth of all cancer-related death per year due to its rapid progression to advanced stages.1 Despite advances in the diagnostic and therapeutic strategies, the prognosis of colon cancer patients has not significantly changed in the past decade.2 Currently, only a few biomarkers, such as mutated KRAS and BRAF, can be used in a preventive strategy to stratify people into appropriate screening, treatment, and prediction programs. Thus, identification of new predictive biomarkers and the underlying molecular mechanisms involved in the progression of colon cancer is urgently needed.
Long noncoding RNAs (lncRNAs), which range from 200 nucleotides to multiple kilobases and lack of protein-coding capacity, are shown to play important roles in diverse physiological and pathological processes, such as development, immune response, metabolism, self-renewal, chemoresistance, and tumorigenesis.3–6 Recently, the regulatory roles of FOXF1 adjacent noncoding developmental regulatory RNA (FENDRR) in human breast cancer, prostate cancer, lung cancer, and gastric cancer are demonstrated.7–9 However, the functions and molecular mechanism of FENDRR in colon cancer have not been investigated.
In this study, the expression of FENDRR, as well as the correlation between FENDRR and its clinicopathological significance in colon cancer, was investigated. FENDRR was identified as a regulator of SOX4 in colon cancer. We found that FENDRR inhibits the progression of colon cancer through repressing the expression of SOX4 protein.
Materials and methods
Tissue specimens and cell culture
The primary cancer tissues and matched adjacent normal mucosa 5 cm distant from the edge of original tumor were obtained from 60 colon cancer patients who underwent surgery without prior chemotherapy or radiotherapy. The diagnosis of every specimen was histopathologically confirmed by two pathologists. The tumor staging was determined according to the TNM staging system of the International Union against Cancer. This study was approved by The Human Research Ethics Committee of Liaocheng People’s Hospital. The written informed consent was collected from all subjects and the study was conducted in accordance with the Declaration of Helsinki.
The human colon mucosa cell line NCM460 and the colon cancer cell lines SW620, SW480, Hct116, RKO and CaCO2 were obtained from the Type Culture Collection of the Chinese Academy of Science (Shanghai, China). All cell lines were cultured in modified Eagle’s medium (Hyclone, Logan, UT, USA) supplemented with 10% FBS (Gibco, Carlsbad, CA, USA) and maintained at 37°C under a humidified atmosphere containing 5% CO2. For plasmid transfection, 1×105 cells/well were seeded in six-well plates overnight and then transfected with plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The levels of FENDRR and SOX4 were confirmed using qRT-PCR or Western blot analysis, respectively.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cultured cells, fresh primary tumors and adjacent normal mucosa of 60 colon cancer patients using TRIzol reagent (Invitrogen). After assessing RNA intensity and purity, cDNA was synthesized from 2 μg total RNA using the PrimeScript RT reagent kit (Takara, Dalian, China). First-strand cDNA was used as a template for qRT-PCR with SYBR Premix Ex Taq (Takara) according to the manufacturer’s instructions. The relative gene expression was calculated by using 2−ΔΔCt method. Primer sequences used in the assays were as follows: FENDRR, forward 5′- CCACA TGGAT GGTTG CCACT CTC-3′ and reverse 5′- GCTGG TACTC GGCCT TCTAA TTGG-3′; SOX4, forward 5′-ACAAT GCCGA GAACA CGGAA GC-3′ and reverse 5′-GATCT GCGAC CACAC CATGA AGG-3′; GAPDH, forward 5′-GGAGC GAGAT CCCTC CAAAA T-3′ and reverse 5′- GGCTG TTGTC ATACT TCTCA TGG-3′.
Western blot analysis
Total protein was extracted from cell lines using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) and the concentration was then quantified by a BCA protein assay kit (Beyotime Biotechnology). Equal amounts of protein (20 μg) were subjected to 10% SDS-PAGE and then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After being blocked in 5% fat-free milk solution containing 0.1% Tween-20 for 1 hr at room temperature, the membranes were then incubated with primary detection antibodies (1:1,000 dilution for E-cadherin, CST, Boston, MA, USA; 1:1,000 dilution for N-cadherin, CST; 1:1,000 dilution for Vimentin, CST; 1:1,000 dilution for SOX4, CST; 1:2,000 dilution for GAPDH, Proteintech, Chicago, IL, USA) at 4°C overnight. After incubation with secondary antibody and washing with 1% Tris-buffered saline Tween (TBST), proteins were detected with ECL detection reagents (Millipore).
Immunohistochemistry
Immunohistochemical staining was performed to examine SOX4 expression in colon cancer tissues. Paraffin-embedded sections were dewaxed and rehydrated and then immersed in boiled citrate buffer (0.01 M, pH 6.0) for antigen retrieval. Staining intensity was scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). Staining area was scored as 0 (no staining of cells), 1 (1–25% positive staining), 2 (26–50% positive staining), 3 (51–75% positive staining), and 4 (>75% positive staining). The staining intensity scores and staining area scores were summed up to give a final total staining scores, based on which the specimens were divided into negative group (0–2), weak group (2–4), moderate group (4–6), and strong group (>6). All the slides were evaluated by two specialists independently who were blinded to patient outcome.
Cell proliferation, colony formation, and invasion assays
<start>For the CCK-8 (Cell Counting Kit-8) assay, transfected cells (SW620 and Hct116) were plated in 96-well plates (2×103 cells/well). At the appropriate time (24, 48, 72, 96 hrs), cell proliferation was measured with a CCK-8 kit (Dojindo Laboratories, Kumamoto-ken, Japan) according to the manufacturer’s instruction. Absorption at 450 nm was measured on a Gen5 microplate reader (BioTek, Winooski, VT, USA).
For the colony formation assay, 1,000 log-phase cells were placed in 6 cm plates and maintained at 37°C under a humidified atmosphere containing 5% CO2, with medium replaced every 3 days. After 14 days, the cells were fixed in methyl alcohol for 30 mins and dyed with Giemsa solution for 60 mins. Visible colonies were then counted and photographed. All assays were performed in triplicate.
Transwell chambers (8-μm pore size, Corning Costar, New York, NY, USA) were used for the invasion assay. 2×105 cells in serum-free medium were placed in the upper compartment of the transwell chamber (Matrigel-coated membrane). Medium containing 10% FBS was added to the lower chamber. After incubation at 37°C in a humidified atmosphere containing 5% CO2 for 48 hrs, cells that invaded through the membrane were fixed and stained with methanol or 0.1% crystal violet, respectively, and then imaged and counted under a microscope. Experiments were performed independently in triplicate.
Confocal immunofluorescent assay
After being first incubated with antibodies specific for E-cadherin (1:1,000, CST), N-cadherin (1:1,000, CST), or Vimentin (1:1,000, CST) for overnight at 4°C, and then with goat anti-rat IgG (1:200, Alexa Fluor 647, Invitrogen) and goat anti-rat IgG (1:200, Alexa Fluor 488, Invitrogen) for 2 hrs at room temperature, cells were mounted by adding DAPI (Southern Biotech, SBA, Birmingham, AL, USA) and examined using a confocal microscope (Olympus, Tokyo, Japan).
Animal studies
The in vivo proliferation assay was carried out with 4-week-old male BALB/C nude mice. Briefly, SW620 cells (1.0×106) stably expressing vector, SOX4 or SOX4 + FENDRR were injected subcutaneously into the groin of nude mice (n=5 for either group). One month after implantation, the mice were killed and tumor weight was measured. All of the animal procedures were conducted following the Animal Care Guidelines of Taishan Medical University. The animal studies were approved by the ethics committee of Taishan Medical University.
Statistical analysis
SPSS 19.0 statistical software package (SPSS Inc, Chicago, IL, USA) was utilized for all data analyses. The differences between the two groups were compared with the Student’s t-test, one-way ANOVAs, or χ2 tests, when appropriate. The correlation between FENDRR levels and SOX4 protein expression was analyzed using Spearman’s correlation coefficient test. Overall survival (OS) curves were estimated by using the Kaplan–Meier method. Ap -value of less than 0.05 was considered statistically significant.
Results
FENDRR expression is significantly downregulated in colon cancer
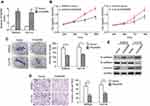
qRT-PCR analysis was performed to evaluate FENDRR expression in 60 paired colon cancer specimens. Results showed that FENDRR transcript was downregulated in 70% (42/60) colon cancer tissues compared with the matched adjacent normal mucosa (p<0.001, Figure 1A). The relationship between FENDRR expression and different clinicopathological features is summarized in Table 1. Attenuation of FENDRR was significantly associated with the pT stage (p=0.022), pN stage (p=0.010), and TNM stage (p<0.001). There was no significant correlation between FENDRR expression and age, gender, or differentiation and vessel invasion. Consistent with our findings, FENDRR was also found to be significantly downregulated in The Cancer Genome Atlas (TCGA) dataset of colon adenocarcinoma (COAD) samples integrated by starBase v3.0 project (
 | Table 1 Association between FENDRR expression and clinicopathological characteristics in colon cancer |
FENDRR overexpression inhibits colon cancer cell proliferation, invasion, and epithelial–mesenchymal transition (EMT)
To explore the possible function of FENDRR in colon cancer cells, we elevated the expression of FENDRR. As a result, overexpression of FENDRR in SW620 and Hct116 cells after treatment with pcDNA3.1-FENDRR was confirmed by qRT-PCR analysis (Figure 2A). The FENDRR-enhanced cells exhibited a significant viability inhibition compared with the control, as measured with a CCK-8 assay (Figure 2B). Colony formation assay revealed that elevation of FENDRR attenuated visible colonies (Figure 2C). Moreover, in the transwell assay, pcDNA3.1-FENDRR impeded the invasive ability of colon cancer cells effectively (Figure 2D). Interestingly, altered FENDRR expression also led to the increased expression of E-cadherin, together with the reduced expression of N-cadherin and Vimentin in colon cancer cells as determined by Western blot analysis (Figure 2E). These findings suggested that FENDRR inhibited the progression of colon cancer in vitro.
FENDRR represses the expression of SOX4 protein in colon cancer
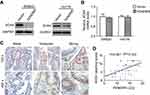
We next sought to explore the mechanisms behind the potent effect of FENDRR on the progression of colon cancer. Recently, many lncRNAs have been confirmed to interact with protein and then increase or decrease the stability of protein.10,11 To identify protein species interacted by FENDRR, we performed a prediction via the RNA association interaction database RAID v2.0 (
FENDRR-mediated SOX4 repression attenuates the progression of colon cancer in vitro
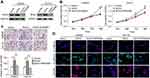
To assess whether the SOX4 stability is a direct mechanism involved in FENDRR-inhibited progression of colon cancer, in vitro assays were performed after SOX4 and FENDRR overexpression (Figure 4A). Notably, forced expression of SOX4 significantly enhanced proliferation and invasive ability of colon cancer cells, and these effects were abolished by activation of FENDRR (Figure 4B and C). On the other hand, via a confocal immunofluorescent assay, the increased levels of mesenchymal markers N-cadherin and Vimentin and decreased expression of epithelial marker E-cadherin were observed both in SW620-SOX4 and in Hct116-SOX4 cells compared with control groups, and restoration of FENDRR abrogated the above EMT process (Figure 4D). These findings clearly indicated that FENDRR-mediated SOX4 repression is probably critical in attenuation of the progression of colon cancer.
FENDRR-mediated SOX4 repression inhibits xenograft tumor growth
To explore the in vivo effect of FENDRR/SOX4 axis on colon cancer cell proliferation, SW620 cells stably expressing vector, SOX4, or SOX4+FENDRR were injected subcutaneously into the groin of nude mice. Results showed that both tumor volume (Figure 5A and B) and weight (Figure 5C) increased in SOX4 overexpression group compared with the control group, and ectopic expression of FENDRR abolished the above effects. These in vivo findings confirmed an important role of FENDRR/SOX4 axis in colon cancer, which was consistent with the in vitro results.
Discussion
In the present study, we documented that downregulation of lncRNA FENDRR was a frequent event in colon cancer tissues and strongly linked to increased risk of poor survival. Furthermore, we provide evidence that low FENDRR expression was significantly correlated with aggressive phenotype of colon cancer cells. FENDRR inhibits the progression of colon cancer at least partly through modulation of SOX4 protein.
FENDRR, also known as lncRNA FOXF1-AS1, is located at chromosomal band 3q13.31 and transcribed from the FOXF1 promoter and co-expressed with FOXF1 gene.14 Khalil et al first identified FENDRR and confirmed its chromatin-modifying complex-binding ability.15 As an important tumor-repressor gene, FENDRR was selected as an optimal biomarker for hepatitis B virus (HBV)-associated hepatocellular carcinoma with great diagnostic and prognostic values.16 FENDRR exerted repressive influences on gastric cancer cell migration and invasion via negative regulation of fibronectin 1 (FN1) and secreted matrix metalloproteinase 2/9 (MMP2/9).17 Moreover, FENDRR depletion facilitates EMT process and metastasis in non-small-cell lung cancer cells.18 Consistent with the previously published reports, attenuation of FENDRR expression both in colon cancer tissues and in cancer cell lines was found in our experiments. We also confirmed that restoration of FENDRR in colon cancer cells coincided with the inhibition of EMT program. It is worth noting that patients with low FENDRR expression appeared to be closed to better clinicopathological features like good differentiation or pT2 stage according to the results shown in Table 1. Considering that our study was conducted in only 60 patients, a larger prospective clinical investigation is warranted to verify the association between FENDRR expression and clinicopathological characteristics in colon cancer.
Accumulating evidence showed that lncRNAs could directly interact with proteins and exert impacts on the stability of proteins. For instance, LncRNA-CYTOR was found to associate with cytoplasmic β-catenin, thus protecting β-catenin from phosphorylation by CK1.19 LncRNA ANCR was shown to bind to EZH2 by stem-loop structure, which probably facilitates the recognition and binding of CDK1 on EZH2 to phosphorylate and finally degrade the protein through the ubiquitin–proteasome pathway.20 It is a remarkable fact that FENDRR was proved to target and repress some nuclear proteins to govern the cells’ state.14,15 Among the nuclear proteins, we selected SOX4 as the research objective because it was reported that SOX4 is required not only for the maintenance of tumorigenesis but also for the initiation of EMT in multiple tumor types.21,22 SOX4 is a member of the group C of Sry-related HMG box transcription factors, and it functions as a higher-order master regulator of EMT by governing the expression of epigenetic modifiers.23 Considering that the interactive correlation was predicted between FENDRR transcript and SOX4 protein, and FENDRR-mediated repression of SOX4 protein levels (rather than mRNA levels) was also observed in our study, we speculated that FENDRR might regulate the progression of colon cancer via the FENDRR/SOX4 axis. In fact, our findings demonstrated that aberrant SOX4 expression contributes to colon cancer cell proliferation, invasion, EMT, and xenograft tumor growth, and these effects were abrogated when FENDRR was upregulated in SOX4-overexpressing cells. All these results suggested that overexpression of FENDRR inhibits the progression of colon cancer at least in part through modulating the SOX4 protein.
Conclusion
Collectively, our findings elucidate a hitherto unexplored FENDRR/SOX4 axis, and it would be interesting and significant to further analyze the FENDRR/SOX4 expression in the same colon cancer samples by in situ hybridization and determine the details of how FENDRR physically interacts with SOX4 in the future research. Taken together, the discovery of FENDRR and its interaction with SOX4 may provide a promising option for facilitating the investigation of colon cancer.
Acknowledgment
We thank Daogui Yang for his excellent technical assistance.
Disclosure
The authors declare that they have no conflicts of interest concerning this article.
References
1. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. doi:10.3322/caac.21220
2. Azvolinsky A. Colorectal cancer: to stack or sequence therapy? J Natl Cancer Inst. 2015;107:5. doi:10.1093/jnci/djv138
3. Ma Y, Yang Y, Wang F, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65(9):1494–1504. doi:10.1136/gutjnl-2014-308392
4. Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi:10.1016/j.ccell.2016.03.004
5. Zhu P, Wu J, Wang Y, et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol. 2018;20(10):1134–1144. doi:10.1038/s41556-018-0194-0
6. Huang D, Chen J, Yang L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19(10):1112–1125. doi:10.1038/s41590-018-0207-y
7. Luo T, Zhao J, Lu Z, et al. Characterization of long non-coding RNAs and MEF2C-AS1 identified as a novel biomarker in diffuse gastric cancer. Transl Oncol. 2018;11(5):1080–1089. doi:10.1016/j.tranon.2018.06.007
8. Li Y, Zhang W, Liu P, et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Onco Targets Ther. 2018;11:1403–1412. doi:10.2147/OTT.S149511
9. Zhang G, Han G, Zhang X, et al. Long non-coding RNA FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. Biomarkers. 2018;23(5):435–445. doi:10.1080/1354750X.2018.1443509
10. Qu L, Wu Z, Li Y, et al. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat Commun. 2016;7:12692. doi:10.1038/ncomms12692
11. Wang P, Xue Y, Han Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–313. doi:10.1126/science.1251456
12. Peng X, Liu G, Peng H, Chen A, Zha L, Wang Z. SOX4 contributes to TGF-beta-induced epithelial-mesenchymal transition and stem cell characteristics of gastric cancer cells. Genes Dis. 2018;5(1):49–61. doi:10.1016/j.gendis.2017.12.005
13. Liu F, Wu L, Wang A, et al. MicroRNA-138 attenuates epithelial-to-mesenchymal transition by targeting SOX4 in clear cell renal cell carcinoma. Am J Transl Res. 2017;9(8):3611–3622.
14. Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi:10.1016/j.devcel.2012.12.012
15. Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi:10.1073/pnas.0904715106
16. Mou Y, Wang D, Xing R, et al. Identification of long noncoding RNAs biomarkers in patients with hepatitis B virus-associated hepatocellular carcinoma. Cancer Biomark. 2018;23(1):95–106. doi:10.3233/CBM-181424
17. Xu TP, Huang MD, Xia R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi:10.1186/s13045-014-0063-7
18. Miao L, Huang Z, Zengli Z, et al. Loss of long noncoding RNA FOXF1-AS1 regulates epithelial-mesenchymal transition, stemness and metastasis of non-small cell lung cancer cells. Oncotarget. 2016;7(42):68339–68349. doi:10.18632/oncotarget.11630
19. Yue B, Liu C, Sun H, et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/beta-catenin signaling promotes metastasis of colon cancer. Molecular Therapy. 2018;26(5):1287–1298. doi:10.1016/j.ymthe.2018.02.024
20. Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24(1):59–71. doi:10.1038/cdd.2016.95
21. Vervoort SJ, Lourenco AR, Tufegdzic Vidakovic A, et al. SOX4 can redirect TGF-beta-mediated SMAD3-transcriptional output in a context-dependent manner to promote tumorigenesis. Nucleic Acids Res. 2018. doi:10.1093/nar/gky755
22. Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi:10.1186/s12943-017-0680-1
23. Tiwari N, Tiwari VK, Waldmeier L, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23(6):768–783. doi:10.1016/j.ccr.2013.04.020
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.