Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA EMX2OS, Regulated by TCF12, Interacts with FUS to Regulate the Proliferation, Migration and Invasion of Prostate Cancer Cells Through the cGMP-PKG Signaling Pathway
Authors Wang Z, Zhang C, Chang J, Tian X, Zhu C, Xu W
Received 24 December 2019
Accepted for publication 11 June 2020
Published 21 July 2020 Volume 2020:13 Pages 7045—7056
DOI https://doi.org/10.2147/OTT.S243552
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Zhiqiang Wang,* Chaowei Zhang,* Junkai Chang, Xin Tian, Chaoyang Zhu, Weibo Xu
Department of Urinary Surgery, Huaihe Hospital of Henan University, Kaifeng 475000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Weibo Xu
Department of Urinary Surgery, Huaihe Hospital of Henan University, No. 8 Baobei Road, Kaifeng 475000, People’s Republic of China
Tel/ Fax +86 371-23906691
Email [email protected]
Background: LncRNA EMX2OS (EMX2 opposite strand/antisense RNA) is notably downregulated in prostate cancer (PCa) tissues and may be regarded as a potential molecular biomarker for diagnosis and prognosis. However, its exact role in regulating the development of PCa is obscure.
Methods: The EMX2OS expression was assessed in PCa tissues, paracancer tissues, PCa cells and normal prostate epithelial cells by qPCR. Gain- and loss-of-function experiments were performed to investigate the role of EMX2OS and FUS in cGMP-PKG (cyclic guanosine monophosphate-dependent protein kinase)-mediated proliferation, invasion, and migration in human PCa cell lines DU145 and PC3. Then, the interaction of transcription factor 12 (TCF12) with EMX2OS promoter was confirmed by using the dual-luciferase reporter and chromatin immunoprecipitation (ChIP) assays. RNA binding protein immunoprecipitation and RNA pull-down assays were used to verify the interaction between EMX2OS and FUS protein. Finally, the role of EMX2OS and FUS in tumor growth in vivo was validated in a xenograft nude mouse model.
Results: TCF12 and EMX2OS were both downregulated in PCa tissues and cells, and they negatively regulated cell proliferation, migration and invasion, and activated cGMP-PKG pathway in DU145 and PC3 cells. TCF12 was a transcription factor of EMX2OS. TCF12 and EMX2OS overexpression both down-regulated cell proliferation, migration and invasion, and activated cGMP-PKG pathway in DU145 and PC3 cells. Furthermore, EMX2OS directly bound with FUS protein and had a synergy effect with FUS protein on cGMP-PKG-mediated cell functions, which could be suppressed by (D)-DT-2 (a cGMP-PKG inhibitor). In addition, the overexpression of FUS or EMX2OS individually markedly decreased the volume and weight of tumors in vivo, and co-overexpression of them further inhibited tumor growth.
Conclusion: EMX2OS, transcriptionally regulated by TCF12, played a synergy role with FUS protein in regulating the proliferation, migration and invasion of PCa cells by activating the cGMP-PKG pathway.
Keywords: LncRNA EMX2OS, TCF12, FUS, cGMP-PKG pathway, prostate cancer
Introduction
Prostate cancer (PCa) is the second most common tumor in men and the second leading cause of cancer death in men, accounting for 27% of cancer patients in Western countries.1 The incidence and mortality of PC among Asian men have increased rapidly over the past few decades.2 Due to the lack of specific and sensitive methods for early PC screening, most patients are diagnosed at advanced stages.3 Therefore, it is necessary to further explore the pathogenesis of PCa and develop therapeutic targets for PCa patients.4
Long non-coding RNAs (lncRNAs) are a cluster of transcripts longer than 200 nucleotides without the ability to encode proteins.5 Increasing evidence suggests that lncRNAs play vital roles in complex physiological and pathological processes, including embryogenesis, organogenesis, and tumorigenesis.6–9 LncRNA EMX2OS is a long-chain non-coding RNA transcribed from the antisense strand of EMX2.10 It has been reported to be involved in some cancers, like papillary thyroid cancer (PTC), endometrial cancer, etc.11,12 However, the role of EMX2OS in the progression of PCa is rarely reported.
Transcription factor 12 (TCF12) belongs to the class I helix-loop-helix (HLH) protein family known as E protein, which can bind to DNA E-box site.13 Increasing studies have shown that TCF12 acts as an oncogene/a tumor suppressor in a variety of human cancers.14 For instance, TCF-12 is a potential molecular target that inhibits the invasion and metastasis of gallbladder cancer cells.15 TCF12 is significantly elevated in cancer-related fibroblasts, causing extracellular matrix remodeling and triggering invasion and metastasis of breast cancer cells in vitro and in vivo.16 In addition, it has been reported that decreasing TCF12 expression contributes to the progression in patients with prostate cancer.17
Protein kinase G (PKG) is a major receptor of the cGMP second messenger. By binding to PKG, cGMP regulates intracellular signaling pathways that control a wide range of intracellular processes, such as cell differentiation, platelet activation, memory formation, and vasodilation.18,19 PKG1 has been identified as a tumor suppressor. cGMP-dependent PKG 2 has been reported to suppress cancer cell proliferation, like glioma cells,20 and promote the apoptosis of breast cancer cells.21
In this study, we found that EMX2OS was lowly expressed both in PCa tissues and cells. It was regulated by TCF12 and had a synergistic effect with FUS protein on cell proliferation, migration, invasion of DU145 and PC3 cells and tumor growth in mice. What is more, EMX2OS activated cGMP-PKG pathway and might play its role by cGMP-PKG pathway.
Materials and Methods
Tissue Sample Collection
The paired tumorous and adjacent normal human prostate tissues were obtained from 25 patients who signed written informed consent after surgery. This study was approved by the Ethics Committee of Huaihe Hospital of Henan University (Kaifeng, China; approval number: HNHHYY-2017-1205).
Cell Culture
LNCaP, DU145, PC3, RWPE-1 and HEK293A cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum and cultured at 37 °C in 5% CO2.
Cell Transfection
We first constructed adenovirus-mediated TCF12, EMX2OS and FUS overexpression vectors. The blank adenoviral vector was used as a negative control. The siRNAs against TCF12, EMX2OS and FUS and negative control siRNAs were synthesized and verified by Thermo Fisher Scientific. All transfections were performed by using Lipofectamine ® 3000 (Thermo, Waltham, MA, USA) according to the manufacturer’s instructions.
Quantitative Polymerase Chain Reaction (qPCR)
Total RNA isolated from cells and tissues using Trizol reagent, according to the manufacturer’s instruction. cDNA templates were generated by reverse transcriptase reactions using SuperScript III reagent (Takara, Dalian, China). RNA abundances were detected by a Fluorescence quantitative PCR reagent kit (Takara, Dalian, China) under the following conditions: 95 °C for 3 min, followed by denaturation at 94 °C for 15 s, annealing at 55 °C for 25 s and extension at 72 °C for 15 s for 35 cycles. The relative differences in mRNA levels were calculated by 2−ΔΔCT method. 18S RNA was regarded as the internal reference. The following primer sequences were used: EMX2OS forward 5ʹ -gtgacttgcacaaggacacaa-3ʹ, reverse 5ʹ -cctgtvtggccattcctct-3ʹ.
Western Blotting
Total protein isolated from cells and tissues using RIPA lysis buffer (ProMab Biotechnology, USA). Equal amounts of protein were separated on 10% SDS polyacrylamide gels and then electrotransferred to polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). After blocking in 5% skim milk, the membranes were incubated overnight at 4 °C with primary antibodies including anti-EMX2OS (1:400; Abcam, Cambridge, UK), anti-TCF12 (1:400; Abcam), anti-FUS (1:400; Abcam), anti-PKG1 (1:500; Cell Signaling Technology, Boston, MA), anti-PKG2 (1:500; Cell Signaling Technology) and β-actin (1:600; Abcam). Next, the membranes were incubated with proper horseradish peroxidase (HRP)-conjugated IgG for 1 h at 37 °C. Protein bands were visualized using an enhanced chemiluminescence detection system (ECL; Beyotime, Shanghai, China).
Cell Counting Kit-8
A Cell counting kit-8 (CCK-8) was used to detect cell proliferation. Cells were seeded into 96-well plates at 1000 cells/well. A 10 μL of CCK-8 reagent was added to each well and incubated for 2 h at 37 °C with 5% CO2. The absorbance was measured at a wavelength of 450 nm.
Transwell Assay
The upper chamber surface of the bottom membrane of the Transwell chamber was coated and placed at 37 °C for 30 min. Cells (2 × 105) were suspended in 500 μL serum-free medium and added to the upper chamber. The lower chamber was supplemented with RPMI-1640 medium containing 10% fetal bovine serum. Membrane infiltrating cells were fixed with 3.7% paraformaldehyde, DAPI stained, and counted under a 400-fold microscope. In addition, cell migration ability was also investigated by the same method, but the upper chambers were not pre-plated by Matrigel. We randomly selected five regions to count the number of invasive and migratory cells and calculate the average value.
Bioinformatics Analysis
CHIPBase (http://rna.sysu.edu.cn/chipbase/) online database was used to analyze the promoter of EMX2OS, and predict the binding sites between TCF12 and EMX2OS. FUS binding motif was provided by the online bioinformatics database StarBase 2.0 (http://starbase.sysu.edu.cn/). The motif was aligned with the EMX2OS sequence.
Luciferase Reporter Assays
Full-length DNA coding sequence of TCF12 was inserted into pGL3-basic vector (USA), and then three vectors containing different inserts cloned from EMX2OS were constructed downstream of TCF12. All constructs were confirmed by DNA sequencing. HEK293A cells were transfected with pGL3-TCF12-EMX2OS reporters. After transfection, we used a luciferase reporter assay (Promega, USA) on a Berthold AutoLumat LB9507 rack luminometer to measure the luciferase activity. The results were expressed as relative luciferase (Luc) activity.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed using ChIP-IT Expression Chromatin Immunoprecipitation Kits (Active motif, Carlsbad, CA, USA). In short, formaldehyde was used to treat cells for 10 min, and Glycine Stop-Fix solution was used to quench the reaction. Next, we achieved 100–500 bp DNA fragments after lysing and sonicating the cells. The immunocomplexes were precipitated by using antibodies against TCF12 and normal serum IgG overnight and rotationally incubation at 4 °C. The 20% of chromatin without antibody incubation was used as the input control. Finally, the reaction mixtures were eluted by using beads according to the manufacturer’s instruction.
Determination of cGMP Accumulation
We used the cGMP ELISA Kit (Sigma, St. Louis, MO, USA) to measure the cGMP accumulation. In brief, 100 μL of standard solution or samples were added to be tested, and mixed gently at 37 °C for 120 min. A 100 μL of test solution was added into each well at 37 °C for 60 min. The plate was washed 3 times. Next, 100 μL of test solution B working solution was added to each well, and the plate was washed 5 times at 37 °C for 60 min. Then, 90 μL substrate solution was added in each well. The reaction was terminated by sequentially adding 5 μL of the stop solution to each well. The optical density (OD value) was measured by an enzyme-coupled instrument at a wavelength of 910 nm.
RNA Binding Protein Immunoprecipitation (RNA-IP) Assay
Cells were washed with ice-cold PBS, and then lysed in Tris/HCl (PH7.5, buffered with 1% Triton containing protease inhibitors), and incubated with 2 μg FUS antibody at 4 °C for 2 h. Next, 40 μL of protein A-Sepharose was added, and the mixture was incubated at 4 °C for 4 h. After centrifugation, the pellets were washed with PBS and resuspended in 0.5 mL Tri Reagent (Sigma-Aldrich). The precipitated RNA eluted in the aqueous solution was analyzed by qPCR.
RNA Pull-Down Assay with Biotinylated RNA Probe
Cells were transfected with 50 nM biotinylated RNA probe for 48 h and then washed with PBS followed by brief vortex, and incubated in a RNA pull-down lysis buffer (Ambion) for 10 min on ice. The lysates were pretreated by centrifugation. A 60 μL (30%) of the samples were separated for input. The remaining lysates were incubated with M-280 streptavidin magnetic beads which were pre-coated with RNase-free BSA and yeast tRNA (Sigma) for 3 h at 4 °C. Then, the beads were washed twice with ice-cold lysis buffer, three times with an SDS-Tris low salt buffer, and once with a high salt buffer. The bound complexes were purified for the subsequent analysis.
In vivo Tumorigenesis Assays
A 1×106 of treated DU145 cells were subcutaneously injected into the right flanks of nude mice. Tumor length (L) and width (W) were measured every 5 days, and tumor volume was calculated using the following equation: volume = (W2 × L)/2. After 40 days, the mice were sacrificed, and tumor volume and weight were measured. This study was performed following the institutional and national guidelines and regulations of the Animal Protection and Use Committee and approved by the Ethics Committee of Huaihe Hospital of Henan University (HNHHYY-2017-1205).
Statistical Analysis
All statistical analyses were carried out using SPSS 22.0, and data were presented as mean ± standard deviation (SD). The differences between the two groups were compared by student’s t-test. ANOVA was used to evaluate the significance of differences in mean values within and between multiple groups. The statistical significance was defined as p<0.05.
Results
EMX2OS Was Lowly Expressed in Prostate Cancer Cell Lines and Tissues
Previous researches have revealed that EMX2OS is differentially expressed in non-prostate and prostate cancer patients,22 but the specific regulatory mechanism of EMX2OS in prostate cancer remains unclear. Therefore, we examined the expression of EMX2OS in prostate cancer tissues and three prostate cancer cell lines (LNCap, DU145 and PC3). The data indicated that EMX2OS was notably low expressed both in prostate cancer tissues and cell lines compared to adjacent normal tissues and normal prostate epithelial cell line (RWPE-1) (Figure 1A and B). The DU145 and PC3 cells were selected for subsequent experiments.
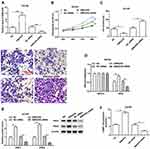
EMX2OS Inhibited Proliferation, Migration and Invasion of DU145 and PC3 Cells and Activated cGMP-PKG Pathway
Next, we investigated the role of EMX2OS played in DU145 and PC3 cells. The cells were transfected with EMX2OS overexpression vector and EMX2OS siRNA (Figure 2A and Supplementary Figure 1A). As shown in results, cell proliferation, migration and invasion were downregulated in DU145 and PC3 cells after overexpressing EMX2OS, whereas inhibiting EMX2OS showed the opposite effect (Figure 2B–D and Supplementary Figure 1B–D). It has demonstrated that EMX2OS is significantly down-regulated in PCa tissues and is closely related to the cGMP-PKG signaling pathway. Our data suggested that the protein levels of PKG1 and PKG2 and the cGMP concentrations in the supernatant were increased in EMX2OS overexpression group, but declined in EMX2OS knockdown group (Figure 2E and F and Supplementary Figure 1 E and F).
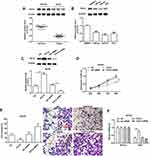
TCF12 Directly Bound to EMX2OS Promoter
Reports have shown that decreasing TCF12 expression contributes to the progression in patients with prostate cancer,17 so there are reasons to suspect a relationship between TCF12 and EMX2OS. ChIPbase online database was used to predict the binding sites between TCF12 and EMX2OS. The potential binding region contained the transcription start site (TSS, chr10: 117,545,068) and a 61 bp length sequence on the intron of EMX2OS (Figure 3A). Then, three luciferase reporters including TCF12 sequence and different truncated sequences of EMX2OS were constructed. Results indicated that the luciferase activities of the reporters containing only the binding region or full-length sequence were markedly increased compared with the control (Figure 3B). In addition, the luciferase activities between the reporter containing non-TCF12 binding region and control had no obvious differences (Figure 3B). Subsequent CHIP assay revealed that EMX2OS promoter expression was observably enriched in the TCF12 antibody-precipitated complex compared to the input, while IgG group did not precipitate the EMX2OS promoter in DU145 and PC3 cells (Figure 3C and Supplementary Figure 2).
Overexpression of TCF12 Inhibited Proliferation, Invasion and Migration of DU145 and PC3 Cells
To further investigate the role of TCF12 played in PCa, we firstly detected the expression of TCF12 in prostate cancer tissues and cell lines. It was found that TCF12 was lowly expressed in tumor tissues and cell lines (LNCap, DU145 and PC3) compared to adjacent normal tissues and RWPE-1 cells (Figure 4A and B). The overexpression of TCF12 prominently impeded the proliferation, migration and invasion of DU145 and PC3 cells (Figure 4C–F and Supplementary Figure 3A–D). After interfering with TCF12, cell proliferation, migration and invasion tended to increase, but not significantly (Figure 4D–F and Supplementary Figure 3A–D).
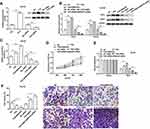
EMX2OS Directly Bound with FUS Protein
It was reported that increased expression of FUS reduced tumor growth and increased apoptosis in prostate cancer cells,23 so we speculated that EMX2OS might bind with FUS protein. StarBase online database was used to search binding motifs of FUS. As shown in Figure 5A, the binding motif of “AACTCT” existed in the EMX2OS sequence (red fonts in Figure 5B). A significant enrichment of EMX2OS was observed in FUS antibody immunoprecipitated protein-RNA complexes by using EMX2OS-specific primers compared with non-specific control primers, but not in that immunoprecipitated by IgG antibody in DU145 and PC3 cells (Figure 5C and Supplementary Figure 4A). The results of RNA pull-down based on EMX2OS-probe displayed that the protein level of FUS in EMX2OS-protein complexes was notably increased along with EMX2OS upregulation (Figure 5D and Supplementary Figure 4B). Furthermore, RNA pull-down assay with WT or MUT biotinylated EMX2OS transcripts revealed that one of the two motifs mutation reduced the binding capacity of EMX2OS with FUS, and both mutations abrogated their binding (Figure 5E and Supplementary Figure 4C).
EMX2OS Played Synergy Effect with FUS Protein in DU145 and PC3 Cells
We further assessed the effects of FUS on cell behaviors and cGMP-PKG pathway in DU145 and PC3 cells. As shown in Figure 6A and Supplementary Figure 5A, we measured the protein level of FUS after transfection with FUS overexpression vector, FUS siRNA and respective controls in DU145 and PC3 cells. The following results revealed that the cell proliferation, migration and invasion were decreased, and PKG1 and PKG2 expression and cGMP concentration were increased after the co-transfection of EMX2OS and FUS, compared with the FUS transfection group (Figure 6B–F and Supplementary Figure 5B–F). The effect of EMX2OS siRNA and FUS siRNA co-treatment on cell proliferation, migration and invasion and cGMP-PKG pathway was stronger than the effect of treatment with FUS siRNA alone (Figure 6B–F and Supplementary Figure 5B–F).
The Effect of FUS and EMX2OS on DU145 and PC3 Cell Behaviors Was Likely to Be Achieved via the cGMP-PKG Pathway
Based on the above tests, we suspected EMX2OS and FUS might play its role via the cGMP-PKG pathway; thus, we used an inhibitor (D)-DT-2 to inhibit the cGMP-PKG pathway (Figure 7A and B and Supplementary Figure 6A and B) in DU145 and PC3 cells. As shown in results, (D)-DT-2 reversed the decrease of cell proliferation, migration and invasion induced by the co-transfection of EMX2OS and FUS overexpression vectors (Figure 7C–E and Supplementary Figure 6C–E).
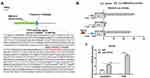
EMX2OS and FUS Synergistically Inhibited Tumor Growth in vivo
Nude mice xenograft assay was performed to assess the effects of EMX2OS and FUS on tumor growth. Our data suggested that the injection of DU145 cells transfected with FUS or EMX2OS overexpression vector significantly inhibited the volume and weight of prostate tumors in vivo compared to the control group (Figure 8A and B). And the co-transfection of FUS and EMX2OS overexpression vectors had a more significant inhibitory effect on tumor volume and weight (Figure 8A and B).
Discussion
Recent evidence suggest that lncRNAs play important regulatory roles in the development of PCa, including cell proliferation, apoptosis, and migration.3 For example, the silencing of lncRNA MalaT1 can inhibit the proliferation, migration and invasion of PCa cells and induce cell cycle arrest.24 LncRNA FEZF1-AS1 promotes cell proliferation and metastasis in PCa via Notch signaling.25 In addition, EMX2OS has been reported to be down-regulated in PTC and endometrial cancer tissues.11,12 In addition, there are reports that EMX2OS is significantly down-regulated in PCa tissues and is closely related to the cGMP-PKG signaling pathway. Our data revealed that EMX2OS was lowly expressed in PCa tissues and cells, and it impeded cell proliferation, migration and invasion, and activated cGMP-PKG pathway in DU145 and PC3 cells. Furthermore, EMX2OS inhibited PCa tumor growth in mice, indicating that EMX2OS was involved in the progression of PCa and it might play a suppressive role in PCa.
Increasing evidence indicate the key role of TCF12 in various human cancers. The aberrant expression of TCF12 has been recently reported to be associated with cancer aggressiveness.26 Functionally, TCF12 acts as an oncogene in gallbladder cancer, breast cancer, and colorectal cancer, but as a tumor suppressor in oral squamous cell carcinoma.13,15,16,27 In addition, decreased expression of TCF12 contributes to progression and predicts biochemical recurrence in patients with PCa.17 Our results demonstrated that TCF12 overexpression prominently impeded the proliferation, migration and invasion of DU145 and PC3 cells. And it was downregulated in cells and tissues of PCa. Suggesting that the abnormally high expression of TCF12 could inhibit the progression of prostate cancer, which was consistent with previous reports.
RNA-binding proteins (RBPs) are key players in post-transcriptional events through its specific and affinitive RNA-binding activities for the sake of the interplay with particular mRNAs to influence cellular activities of diverse tumors.28–30 FUS RNA binding protein (FUS) is a member of a protein cell group called FET/TET. Researches displayed that PCa patients with higher FUS levels have the chance of more survival.23 Furthermore, increased expression of FUS reduces tumor growth and prevents the G1 stage and increases apoptosis in PCa cells.23 In this study, we found that EMX2OS directly bound with FUS protein, and played synergy effect with FUS protein: the cell proliferation, migration, invasion and tumor growth were decreased, and PKG1 and PKG2 expression and cGMP concentration were increased after the co-transfection of EMX2OS and FUS, compared with the FUS transfection group. The effect of EMX2OS siRNA and FUS siRNA co-treatment on cell proliferation, migration, invasion, cGMP-PKG pathway and tumor growth was stronger than the effect of FUS siRNA transfection.
cGMP-dependent signaling pathways have been reported to participate in the biological functions of tumor cells,31 and PKG1 has been identified as a tumor suppressor.32 It has been reported that PKG2 is down-regulated in various human cancer cell lines, including A549 lung, HepG2 liver, OS-RC-2 kidney, SW480 colon cancer and U251 glioma cell lines.21 In addition, cGMP‑dependent PKG II was demonstrated to inhibit the proliferation and migration, and enhance the apoptosis of gastric cancer cells.33 We had testified that cGMP-PKG signaling pathway was strongly associated with the progression of PCa, and EMX2OS activated this pathway and might suppress cell proliferation, migration and invasion of DU145 and PC3 cells via this pathway.
In conclusion, these findings demonstrated that EMX2OS inhibited cell proliferation, migration, invasion of DU145 and PC3 cells and PCa tumor growth in mice, and activated cGMP-PKG pathway. And it was regulated by TCF12, and played synergy effect with FUS protein. Moreover, EMX2OS might play its role by cGMP-PKG pathway.
Acknowledgments
This study was supported by the Scientific and Technological Project of Henan Province in China (No. 192102310109).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. Ito K. Prostate cancer in Asian men. Nat Rev Urol. 2014;11(4):197–212. doi:10.1038/nrurol.2014.42
3. Zhang C, Wang G, Fu B, et al. LncRNA CASC15 promotes migration and invasion in prostate cancer via targeting miR-200a-3p. Eur Rev Med Pharmacol Sci. 2019;23(19):8303–8309. doi:10.26355/eurrev_201910_19141
4. Wang ZY, Duan Y, Wang P. SP1‐mediated upregulation of lncRNA SNHG4 functions as a ceRNA for miR‐377 to facilitate prostate cancer progression through regulation of ZIC5. J Cell Physiol. 2019;1–12. doi:10.1002/jcp.29285
5. Jiang Z, Zhang Y, Chen X, Wu P, Chen D. Long noncoding RNA RBMS3-AS3 acts as a microRNA-4534 sponge to inhibit the progression of prostate cancer by upregulating VASH1. Gene Ther. 2019;1–14. doi:10.1038/s41434-019-0108-1
6. Wo Q, Zhang D, Hu L, et al. Long noncoding RNA SOX2-OT facilitates prostate cancer cell proliferation and migration via miR-369-3p/CFL2 axis. Biochem Biophys Res Commun. 2019;520(3):586–593. doi:10.1016/j.bbrc.2019.09.108
7. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509. doi:10.1007/s00018-016-2174-5
8. Perry BT, Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development. 2016;143(21):3882–3894. doi:10.1242/dev.140962
9. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253. doi:10.1038/nm.3981
10. Yang C-A, Bauer S, Ho Y-C, et al. The expression signature of very long non-coding RNA in myalgic encephalomyelitis/chronic fatigue syndrome. J Transl Med. 2018;16(1):231–238. doi:10.1186/s12967-018-1600-x
11. Gu Y, Feng C, Liu T, Zhang B, Yang L, Ahmad A. The downregulation of lncRNA EMX2OS might independently predict shorter recurrence-free survival of classical papillary thyroid cancer. PLoS One. 2018;13(12):e0209338. doi:10.1371/journal.pone.0209338
12. Noonan FC, Goodfellow PJ, Staloch LJ, Mutch DG, Simon TC. Antisense transcripts at the EMX2 locus in human and mouse. Genomics. 2003;81(1):58–66. doi:10.1016/s0888-7543(02)00023-x
13. Chen YF, Yang CC, Kao SY, et al. MicroRNA-211 enhances the oncogenicity of carcinogen-induced oral carcinoma by repressing TCF12 and increasing antioxidant activity. Cancer Res. 2016;76(16):4872–4886. doi:10.1158/0008-5472
14. Wang X, Gao S, Xie F, et al. High expression of TCF12 contributes to gastric cancer development via being target regulated by miR-183 and activating PI3K/AKT pathway. J Cell Biochem. 2019;120(8):13903–13911. doi:10.1002/jcb.28664
15. He J, Shen S, Lu W, Zhou Y, Shao Y. HDAC1 promoted migration and invasion binding with TCF12 by promote EMT progress in gallbladder cancer. Oncotarget. 2016;7(22):32754–32764. doi:10.18632/oncotarget.8740
16. Tang X, Hou Y, Yang G, et al. Stromal miR-200s contribute to breast cancer cell invasion through CAF activation and ECM remodeling. Cell Death Differ. 2016;23(1):132–145. doi:10.1038/cdd.2015.78
17. Chen QB, Liang YK, Zhang YQ, et al. Decreased expression of TCF12 contributes to progression and predicts biochemical recurrence in patients with prostate cancer. Tumour Biol. 2017;39(6):1–8. doi:10.1177/1010428317703924
18. Osborne B, Wu J, McFarland C, et al. Crystal structure of cGMP-dependent protein kinase reveals novel site of interchain communication. BMC Pharmacol. 2011;19(9):1317–1327. doi:10.1016/j.str.2011.06.012
19. VanSchouwen B, Selvaratnam R, Giri R, et al. Mechanism of cAMP partial agonism in protein kinase G (PKG). J Biol Chem. 2015;290(48):28631–28641. doi:10.1074/jbc.M115.685305
20. Wang R, Kwon IK, Thangaraju M, et al. Type 2 cGMP-dependent protein kinase regulates proliferation and differentiation in the colonic mucosa. Ajp Gastrointest Liver Physiol. 2012;303(2):G209–G219. doi:10.1152/ajpgi.00500.2011
21. Lan T, Chen Y, Sang J, et al. Type II cGMP-dependent protein kinase inhibits EGF-induced MAPK/JNK signal transduction in breast cancer cells. Oncol Rep. 2012;27(6):2039–2044. doi:10.3892/or.2012.1726
22. Xu N, Wu YP, Yin HB, Xue XY, Gou X. Molecular network-based identification of competing endogenous RNAs and mRNA signatures that predict survival in prostate cancer. J Transl Med. 2018;16(1):274. doi:10.1186/s12967-018-1637-x
23. Ghanbarpanah E, Kohanpour MA, Hosseini-Beheshti F, Yari L, Keshvari M. Structure and function of FUS gene in prostate cancer. Bratisl Med J. 2018;119(10):660–663. doi:10.4149/bll_2018_118
24. Dai X, Liang Z, Liu L, et al. Silencing of MALAT1 inhibits migration and invasion by sponging miR13p in prostate cancer cells. Mol Med Rep. 2019;20(4):3499–3508. doi:10.3892/mmr.2019.10602
25. Zhu L, Song L, Xu Q, Zhan J. Highly expressed long non-coding RNA FEZF1-AS1 promotes cells proliferation and metastasis through notch signaling in prostate cancer. Eur Rev Med Pharmacol Sci. 2019;23(12):5122–5132. doi:10.26355/eurrev_201906_18176
26. Shu L, Zhang Z, Cai Y. MicroRNA-204 inhibits cell migration and invasion in human cervical cancer by regulating transcription factor 12. Oncol Lett. 2018;15(1):161–166. doi:10.3892/ol.2017.7343
27. Lee CC, Chen WS, Chen CC, et al. TCF12 protein functions as transcriptional repressor of E-cadherin, and its overexpression is correlated with metastasis of colorectal cancer. J Biol Chem. 2012;287(4):2798–2809. doi:10.1074/jbc.M111.258947
28. Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3:506–528. doi:10.1016/j.trecan.2017.05.003
29. Fagoonee S, Picco G, Orso F, et al. The RNA-binding protein ESRP1 promotes human colorectal cancer progression. Oncotarget. 2017;8(6):10007–10024. doi:10.18632/oncotarget.14318
30. Wu Q, Ma J, Meng W, Hui P. DLX6-AS1 promotes cell proliferation, migration and EMT of gastric cancer through FUS-regulated MAP4K1. Cancer Biol Ther. 2019;1:1–9. doi:10.1080/15384047.2019.1647050
31. Hofmann F. The biology of cyclic GMP-dependent protein kinases. J Biol Chem. 2005;280(1):1–4. doi:10.1074/jbc.R400035200
32. Karakhanova S, Golovastova M, Philippov PP, Werner J, Bazhin AV. Interlude of cGMP and cGMP/protein kinase G type 1 in pancreatic adenocarcinoma cells. Pancreas. 2014;43(5):784–794. doi:10.1097/MPA.0000000000000104
33. Jiang L, Lan T, Chen Y, et al. PKG II inhibits EGF/EGFR-induced migration of gastric cancer cells. PLoS One. 2013;8(4):e61674. doi:10.1371/journal.pone.0061674
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.