Back to Journals » Cancer Management and Research » Volume 12
LncRNA DSCAM-AS1 Promotes Colon Cancer Cells Proliferation and Migration via Regulating the miR-204/SOX4 Axis
Authors Lu C, Xie T, Guo X, Wu D, Li S, Li X, Lu Y, Wang X
Received 20 February 2020
Accepted for publication 13 May 2020
Published 9 June 2020 Volume 2020:12 Pages 4347—4356
DOI https://doi.org/10.2147/CMAR.S250670
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Canrong Lu,* Tianyu Xie,* Xin Guo, Di Wu, Shuo Li, Xiongguang Li, Yixun Lu, Xinxin Wang
Department of General Surgery, Chinese People’s Liberation Army General Hospital, Beijing 100853, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xinxin Wang
Department of General Surgery, Chinese People’s Liberation Army General Hospital, Beijing 100853, People’s Republic of China
Email [email protected]
Introduction: Long non-coding RNA (lncRNA) DSCAM-AS1 was reported to be aberrantly expressed and play pivotal roles in various human cancers. The aim of the present study was to investigate the expression and roles of DSCAM-AS1 in colon cancer (CC).
Methods: Quantitative real-time PCR (qRT-PCR) was used to detect the expression of DSCAM-AS1, miR-204 and the mRNA level of SOX4. Cell proliferation and cell cycle were analyzed by MTT assay and flow cytometry, respectively. Transwell assay was used for migration capacity detection. Luciferase activity assay was conducted to verify the direct binding of DSCAM-AS1 and miR-204 or miR-204 and SOX4. The protein expression of SOX4 was determined by Western blot. Kaplan–Meier curves were calculated and the Log rank test was performed for the survival data analysis.
Results: DSCAM-AS1 was significantly upregulated in CC and high expression of DSCAM-AS1 was associated with poor prognosis in patients with colon cancer. Knockdown of DSCAM-AS1 significantly suppressed CC cells proliferation and migration. In addition, DSCAM-AS acted as a molecular sponge for miR-204 and SOX4 was identified as a direct target of miR-204 in CC. Moreover, the rescue assay revealed that miR-204 inhibition partly abolished the effects of DSCAM-AS1 knockdown on CC cells proliferation, migration and SOX4 expression.
Discussion: The present study demonstrated that DSCAM-AS1 acted as an oncogenic lncRNA in CC progression by regulating miR-204/SOX4 axis and DSCAM-AS1 may serve as a novel therapeutic target in the treatment of colon cancer.
Keywords: DSCAM-AS1, miR-204/SOX4, colon cancer, proliferation, migration
Introduction
Colon cancer (CC), derived from digestive tract, is the third most common malignant tumor and one of the leading causes of cancer-related deaths among men and women.1 What is worse, the incidence and mortality rate of CC is still increasing each year. It was estimated that there will be about 2 million new cases and nearly 1 million deaths of CC occur one year by 2030.2 At present, surgical resection is the most important choice for colon cancer treatment, supplemented by radiotherapy, chemotherapy, and immunotherapy.3,4 Although a lot of advances in the early diagnosis and treatment methods of CC have been achieved during the past few decades, the prognosis of CC patients is still not satisfactory.5 Therefore, it is necessary to identify some novel molecule involved in the initiation and development of colon cancer, so as to develop some new therapeutic methods for CC patients.
Long non-coding RNAs (LncRNAs) are a subgroup of poorly conserved endogenous RNAs containing more than 200 nucleotides in length and have little protein coding ability.6 LncRNAs always play their roles by regulating the RNA and/or the protein content of a cell on the transcriptional and the post-transcriptional levels.7 Accumulating evidence showed that LncRNAs may act as tumor suppressors or oncogenes by participating in multiple processes of cancer development, such as cell proliferation, apoptosis, migration and invasion.8,9 LncRNA Down Syndrome Cell Adhesion Molecule antisense (DSCAM-AS1), located on 21q22.2, is belonged to the immunoglobulin superfamily of cell adhesion molecules.10 Recently, emerging literatures reported that DSCAM-AS1 play crucial roles in various human cancers. For example, Liao et al showed that DSCAM-AS1 was upregulated in non-small cell lung cancer (NSCLC). Its overexpression could enhance NSCLC cell migration and invasion via upregulating BCL11A.11 Huang et al reported that DSCAM-AS1 was significantly upregulated in melanoma. Knockdown of DSCAM-AS1 inhibited proliferation, colony formation, migration and invasion of melanoma cells.12 Ji and colleagues demonstrated that DSCAM-AS1 promoted the progression of hepatocellular carcinoma (HCC) via sponging miR-338-3p and increased DSCAM-AS1 was associated with poor prognosis in HCC.13 However, the expression level and functions of DSCAM-AS1 in colon cancer are still not fully understood.
MicroRNAs (miRNAs) are a class of single-stranded small molecule RNAs that regulate the expression of post-transcriptional genes via binding to its 3ʹuntranslated region (3ʹ-UTR).14 miR-204 was reported to act as a tumor suppressor in multiple human cancers such as glioblastoma, hepatocellular cancer and bladder cancer.15–17 Moreover, it was demonstrated that SOX4 was a target gene of miR-204 and the miR-204/SOX4 axis played important roles in cancers development. For example, Hu et al showed that miR-204 inhibited cell metastasis and EMT in lung adenocarcinoma through targeting SOX4.18 Yin et al demonstrated that miR-204 negatively regulates SOX4 and inhibited proliferation, migration and invasion of T-cell acute lymphoblastic leukemia cell lines.19 Wu et al reported that miR-204 inhibited cell proliferation, migration and invasion in human renal cell carcinoma cells by downregulating SOX4.20 In colon cancer, previous studies showed that SOX4 was overexpressed and associate with a relative poor prognosis.21,22 However, whether lncRNA DSCAM-AS1 play its roles in cancer progression via regulating the miR-204/SOX4 axis is unknown.
In the current study, we aimed to investigate the expression pattern and roles of DSCAM-AS1 in colon cancer. Firstly, we detected the expression levels of DSCAM-AS1 in colon cancer tissues and cell lines in comparison with normal tissues and cell line. Then, we determined the biological functions of DSCAM-AS1 in colon cancer cells by in vitro assays. Finally, we demonstrated that DSCAM-AS1 exerted its effects in colon cancer via regulating the miR-204/SOX4 axis. These results suggest that LncRNA DSCAM-AS1 may serve as a novel biomarker or therapeutic target in colon cancer.
Materials and Methods
Clinical Tissue Samples
In this study, the colon cancer tissues and paired adjacent normal tissues were collected from 37 primary colon cancer patients who underwent surgical treatment at Department of General Surgery, Chinese People’s Liberation Army General Hospital. The clinicopathological characteristics of CC patients were summarized in Table 1. Patients received no chemotherapy or radiotherapy prior surgery. After surgical resection, tissue samples were all frozen in liquid nitrogen immediately and stored at −80°C until use. All the patients enrolled were followed up for at least 60 months and the survival time was recorded. This study was approved by the Ethics Committee of Chinese People’s Liberation Army General Hospital and each patient has signed a written informed consent for all the procedures.
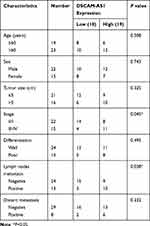 |
Table 1 Correlation Between DSCAM-AS1 Expression and Clinicopathological Characteristics in Colon Cancer Patients |
Cell Culture and Transfection
The human colon cancer cell lines (HT29, HCT8, SW480, and LOVO) and the normal human colon epithelial NCM460 cell line were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were all cultured in Dulbecco’s Modified Eagle medium (DMEM, Hyclone) containing 10% FBS (Gibco), 100 U/mL penicillin and 100 mg/mL streptomycin (Corning). All cell lines were cultured in an incubator at 37°C with 5% CO2.
Cell transfections were all performed using Lipofectamine® 2000 reagent (Invitrogen) according to the manufacturer’s protocol. Briefly, after seeded in 6-well plates and grown to a cell density of 30%, the cells were transfected and cultured at 37°C for further 48 hours. Then, the cells were harvested for qRT-PCR and other experiments. The siRNA targeting lncRNA DSCAM-AS1 (si-DSCAM-AS1) and the negative control (si-NC) were purchased from GenePharma Co. (Shanghai, China). The miRNA mimics, inhibitors and their negative controls were all provided by RiboBio Co. (Guangzhou, China).
RNA Extraction and qRT-PCR
Total RNA was extracted from clinical tissue samples or cultured cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. RNA was reverse transcribed into cDNA using the SuperScript III Reverse Transcriptase kit (Thermo Fisher Scientific, Inc.) and qRT-PCR was performed using the SYBR Green PCR Master mix (Thermo Fisher Scientific, Inc.) according to manufacturer’s protocol. β-actin was used as an internal control for DSCAM-AS1 and SOX4 mRNA levels. U6 was used as the internal control for miR-204. The primers were shown as follow: lncRNA DSCAM-AS1 forward, 5ʹ-GTGACACAGCAAGACTCCCT-3ʹ and reverse, 5ʹ- GATCCGTCGTCCATCTCTGT-3ʹ; β-actin forward, 5ʹ-GGCCCAGAATGCAGTTCGCCTT-3ʹ and reverse, 5ʹ- AATGGCACCCTGCTCACGCA-3ʹ; miR-204 forward, 5ʹ-GACGCTTTCCCTTTGTCATCCT-3ʹ and reverse, 5ʹ-GTGCAGGGTCCGAGGTATTC-3ʹ; U6 forward, 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ and reverse, 5ʹ-GGAACGCTTCACGAATTTG-3ʹ; SOX4 forward, 5ʹ-CTTGACATGATTAGCTGGCATGATT-3ʹ and reverse, 5ʹ-CCTGTGCAATATGCCGTGTAGA-3ʹ. The relative expression levels were normalized with the internal controls and calculated according to the 2− ΔΔCT method.
MTT Assays
Cell proliferation was determined by using (3-(4,5-dimethyl-thiazol-2-y1) 2,5-diphenyl tetrazolium bromide) (MTT) assay. In brief, the transfected cells were seeded into 96-well plates at a density of 5 × 103 cells/well and cultured at 37°C. Following incubation for 24, 48, 72 or 96 hours, 100 μL full culture medium containing 0.5 mg/mL MTT (Sigma-Aldrich, St. Louis, MO, USA) was added into each well and cells were cultured for additional 4 hours. Subsequently, the medium was removed and 150 μL DMSO (Sigma-Aldrich) was added to dissolve the formazan crystals. The absorbance was measured at 490 nm using a microplate reader (Molecular Devices, USA).
Cell Cycle Analysis
Cell cycle distribution was detected by propidium iodide (PI) staining. In short, the transfected cells were collected and fixed with 70% ethanol overnight at 4°C. Then, the cells were washed twice and stained with PI (Sigma-Aldrich) following the manufacturer’s protocol. Cell cycle distribution was determined by using a BD FACS Calibur Flow Cytometer (BD Biosciences, CA, USA).
Transwell Migration Assay
Transwell Chambers (8.0μm pore, BD Biosciences, CA, USA) were used to measures cell migration capacity. In brief, 2 x 105 cells were isolated, resuspended in 200μL serum-free medium and seeded into the upper chambers. The lower chambers were supplemented with 600μL complete medium with 10% FBS. After incubated at 37°C for 24 h, cells on the upper side of the membrane were wiped off by a cotton swab and the migrated cells were fixed with 4% paraformaldehyde for 20 min. Thereafter, the migrated cells were stained with 0.1% crystal violet for 5 min at room temperature and washed with PBS 3 times. The images were photographed using a microscope (Olympus, Tokyo, Japan), and the migrated cells were counted in three random fields.
Luciferase Reporter Assay
Bioinformatics analysis indicated that miR-204 was identified as a candidate target of DSCAM-AS1 by Starbase and SOX4 was identified a direct target of miR-204 by TargetScan. For luciferase reporter assay, the wild type or mutant 3ʹ-UTR of DSCAM-AS1 or SOX4 containing the miR-204 binding site was cloned into the firefly luciferase-expressing pmirGLO vector (Promega, Madison, WI, USA) to generate the wild type or mutant 3ʹ-UTR reporters, named DSCAM-AS1 WT, SOX4 WT, DSCAM-AS1 Mut or SOX4 Mut respectively. The wild type (WT) or mutant (Mut) luciferase vectors were co-transfected with miR-NC or miR-204 mimics into colon cancer cells using Lipofectamine® 2000 according to the manufacturer’s protocol. After 48 hours, the luciferase activity was assessed by using a Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) and normalized to Renilla luciferase activity.
Western Blot
Total protein was extracted from cultured cells using radioimmunoprecipitation assay (RIPA) and then quantified by bicinchoninic acid (BCA) method (Pierce, Rockford, IL, USA). Equal amount of protein sample was separated using 9–11% SDS-PAGE and then transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat milk in PBS for 1 hour at room temperature and incubated with primary antibody (both from Abcam, Cambridge, UK) at 4 °C overnight. After washed three times, membranes were incubated with horseradish peroxidase-labeled secondary antibody (Abcam, Cambridge, UK) for 1 h. Subsequently, the signals were detected using an enhanced chemiluminescent (ECL) Western blot analysis kit (Pierce, Rockford, IL).
Statistical Analysis
Data were expressed as mean ± SD and each experiment was repeated at least three times. All statistical analyses were performed by using GraphPad Prism 5.0 software (San Diego, CA). The comparisons between groups were compared by using the t-test or one-way analysis of variance. The correlation between lncRNA DSCAM-AS1 expression and clinicopathological characteristics of CC patients was analyzed by the Chi-square test. Kaplan–Meier curves were calculated and the Log rank test was performed for the survival data analysis. The potential linear relationship of DSCAM-AS1 and miR-204 expression level was determined by Spearman correlation analysis. The difference was significant when P < 0.05.
Results
DSCAM-AS1 Was Overexpressed and Associated with Poor Prognosis in CC
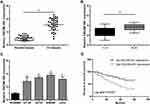
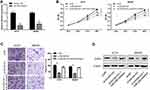
The relative expression level of lncRNA DSCAM-AS1 was detected in CC tissues and cell lines by qRT-PCR. Data revealed that DSCAM-AS1 was significantly overexpressed in CC tissues in comparison with matched adjacent normal tissues (Figure 1A). Meanwhile, we observed that DSCAM-AS1 expression was remarkably higher in CC tissues with advanced clinical stage (III–IV phase) than that with early clinical stage (I–II phase) (Figure 1B). Moreover, our results of qRT-PCR showed that the expression of DSCAM-AS1 was significantly increased in CC cell lines compared with normal human colon epithelial cell line NCM460 (Figure 1C). To evaluate the clinical significance of DSCAM-AS1 in CC, patients were divided into two groups according to the median expression: low DSCAM-AS1 expression and high DSCAM-AS1 expression group. The correlation between DSCAM-AS1 expression and the clinicopathological characteristics of CC patients was evaluated. Data revealed that the expression of DSCAM-AS1 was closely correlated with clinical tumor stage and lymph node metastasis in CC patients (Table 1). In addition, Kaplan–Meier analysis showed that patients with high DSCAM-AS1 expression have a relative poor overall survival (Figure 1D). These results suggest that DSCAM-AS1 may play important roles in the development of CC.
DSCAM-AS1 Knockdown Suppressed CC Cells Proliferation and Migration
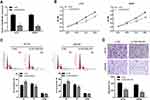
To explore the biological effects of DSCAM-AS1 on CC cells proliferation and migration, the expression of DSCAM-AS1 was effectively knocked down in CC cells HCT8 and SW480 by DSCAM-AS1 siRNAs transfection (Figure 2A). Then, functional assays were conducted on transfected cells. Results of MTT assay showed that knockdown of DSCAM-AS1 significantly suppressed HCT8 and SW480 cells proliferation (Figure 2B). Flow cytometry was used to detect cell cycle distribution of CC cells after DSCAM-AS1 knockdown. Data showed that DSCAM-AS1 knockdown remarkably inhibited cell cycle transition in HCT8 and SW480 cells (Figure 2C). In addition, the migration capacity of transfected CC cells was determined by transwell migration assay, and results revealed that knockdown of DSCAM-AS1 significantly reduced CC cells migration capacity (Figure 2D). These results suggest that DSCAM-AS1 has a significant effect on the progression of colon cancer.
DSCAM-AS1 Acted as a Molecular Sponge for miR-204 in CC Cells
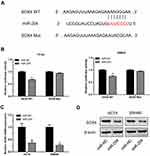
Accumulating studies reported that lncRNAs always exert their functions via acting as competing endogenous RNAs (ceRNA) for some microRNAs in cancer development.23,24 We supposed that DSCAM-AS1 may also play its roles by sponging some microRNA in CC. Bioinformatic analysis by Starbase2.0 (http://starbase.sysu.edu.cn/) revealed that miR-204 contains the potential binding site for DSCAM-AS1 (Figure 3A). Then, the luciferase reporter assay was conducted in HCT8 and SW480 cells to confirm that. Data showed that miR-204 significantly suppressed the luciferase activity of the wild type DSCAM-AS1 reporters (DSCAM-AS1 WT) but not of the mutant reporters (DSCAM-AS1 Mut) in CC cells (Figure 3B). Meanwhile, we found that DSCAM-AS1 expression was obviously reduced following miR-204 overexpression and knockdown of DSCAM-AS1 significantly upregulated miR-204 expression in CC cells (Figure 3C, D). Moreover, the relative expression levels of miR-204 in CC tissues and cell lines were also detected. The results showed that miR-204 was significantly downregulated in CC tissues compared with adjacent normal tissues (Figure 3E). CC cell lines also showed a relative lower expression level of miR-204 than that of the normal colon epithelial cell line NCM460 (Figure 3F). In addition, the relationship between DSCAM-AS1 and miR-204 expression level in CC tissue samples was also evaluated. We found that DSCAM-AS1 expression was negatively correlated with miR-204 level in CC (Figure 3G). These results indicated that DSCAM-AS1 may act as a molecular sponge for miR-204 in CC cells.
SOX4 Was a Direct Target of miR-204 in CC
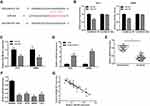
Our results indicated that miR-204 was downregulated in CC tissues and cell lines. Here, we explored the downstream target of miR-204 in CC. We used TargetScan tool to predict the candidate target of miR-204 and found that SOX4 was a direct target of miR-204 (Figure 4A). To confirm that, a luciferase activity assay was conducted in CC cells. Results of luciferase activity assay showed the luciferase activity was significantly reduced following cotransfection of miR-204 mimics and wild-type SOX4 vectors (SOX4 WT), but no change following cotransfection of miR-204 mimics and mutant SOX4 vectors (SOX4 Mut) (Figure 4B). Then, the mRNA and protein expression of SOX4 in CC cells were determined following miR-204 overexpression. Results of qRT-PCR showed that the mRNA level of SOX4 was significantly reduced after overexpression of miR-204 in HCT8 and SW480 cells (Figure 4C). Data from Western blot revealed that the protein expression of SOX4 was also dramatically decreased after miR-204 overexpression in CC cells (Figure 4D). These results suggest that SOX4 was a direct target of miR-204 in CC.
DSCAM-AS1 Exerted Its Effects on CC Cells via miR-204/SOX4 Axis
The results above suggest that DSCAM-AS1 acted as a molecular sponge for miR-204 and SOX4 was a direct target of miR-204 in CC. Previous study reported that SOX4 was overexpressed and associate with a relative poor prognosis in colon cancer.21,22 We supposed that DSCAM-AS1 may perform its effects on CC cells via miR-204/SOX4 axis. To verify it, we transfected miR-204 inhibitors to downregulate its expression in CC cells following DSCAM-AS1 knockdown (Figure 5A). MTT assay indicated that inhibiting of miR-204 partially reversed the suppressive effects of DSCAM-AS1 knockdown on CC cells proliferation (Figure 5B). The results of migration assay also indicated that the effects of DSCAM-AS1 knockdown on CC cells migration capacity was partly reversed (Figure 5C). Furthermore, we detected the protein expression of SOX4 in the treated cells. Data of Western blot showed that the protein expression of SOX4 was significantly reduced after knockdown of DSCAM-AS1, while this effect was partly restored when co-transfected with miR-204 inhibitors (Figure 5D). These results suggest that DSCAM-AS1 exerted its effects on CC cells via regulating miR-204/SOX4 axis.
Discussion
An increasing number of studies have reported that the dysregulation of lncRNAs are involved in the tumorigenesis and development of multiple human cancers including colon cancer. For examples, it was reported that lncRNA UCA1 was upregulated in colon cancer and could modulate progression of colon cancer through regulating the miR-28-5p/HOXB3 axis.25 LncRNA FENDRR was demonstrated to function as a tumor-suppressor gene by repressing SOX4 and may serve as a potential therapeutic target in colon cancer.22 Besides, lncRNA HOTAIR was showed to be overexpressed in colon cancer and promote malignant progression of colon cancer by regulating miR-34a.26 However, the clinical significance and biological effects of lncRNA DSCAM-AS1 in colon cancer have not full investigated.
In this study, we are aimed to investigate the expression pattern and functions of DSCAM-AS1 in colon cancer. Firstly, we used qRT-PCR to detect the expression level of DSCAM-AS1 in colon cancer. We found that DSCAM-AS1 was significantly upregulated in CC tissue samples and cell lines when compared with normal control. We also observed that the expression of DSCAM-AS1 was closely correlated with clinical tumor stage and lymph node metastasis in CC patients. Kaplan–Meier analysis showed that patients with high expression of DSCAM-AS1 have a significantly shorter overall survival time. Then, we transfected siRNAs to downregulate the expression of DSCAM-AS1 in CC cells and performed some functional assays. Our results showed that knockdown of DSCAM-AS1 significantly suppressed CC cells proliferation and cell cycle transition. Moreover, the transwell migration assay indicated that the migration capacity of CC cells was significantly reduced following DSCAM-AS1 knockdown. These results suggest that DSCAM-AS1 was upregulated and acted as an oncogene in colon cancer.
During the past few years, accumulating evidence suggest that lncRNAs can regulate the progression of human cancers by acting as competing sponges for some miRNAs. Here, it was predicted that miR-204 contains the potential binding site for DSCAM-AS1 by Starbase2.0. We further demonstrated that DSCAM-AS1 acted as s molecular sponge for miR-204 in CC cells and SOX4 was identified as a direct target of miR-204 in CC. These results revealed that DSCAM-AS1 may play its roles in CC by regulating miR-204/SOX4 axis. To verify this, a rescue assay was performed on CC cells. We observed that miR-204 inhibition partially reversed the suppressive effects of DSCAM-AS1 knockdown on CC cells proliferation and migration. Results of Western blot showed that the protein levels of SOX4 were reduced after DSCAM-AS1 knockdown and these effects were partly abolished by miR-204 inhibition. These results suggest that DSCAM-AS1 may play its roles via regulating miR-204/SOX4 axis in colon cancer.
Taken together, the present study demonstrated that DSCAM-AS1 was upregulated in CC and associated with a poor prognosis. DSCAM-AS1 acted as an oncogenic lncRNA in CC progression by regulating miR-204/SOX4 axis. These results suggest that DSCAM-AS1 may serve as a novel therapeutic target in the treatment of colon cancer.
Conclusions
The present study demonstrated that lncRNA DSCAM-AS1 was upregulated and associated with a poor prognosis in CC. DSCAM-AS1 knockdown suppressed CC cells proliferation and migration by regulating miR-204/SOX4 axis. DSCAM-AS1 may serve as a novel therapeutic target in the treatment of colon cancer.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declared no conflicts of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Xiong W, Qin J, Cai X, et al. Overexpression LINC01082 suppresses the proliferation, migration and invasion of colon cancer. Mol Cell Biochem. 2019;462(1–2):33–40. doi:10.1007/s11010-019-03607-7
3. Xi X, Teng M, Zhang L, Xia L, Chen J, Cui Z. MicroRNA-204-3p represses colon cancer cells proliferation, migration, and invasion by targeting HMGA2. J Cell Physiol. 2019.
4. Ma Y, Guan L, Han Y, et al. siPRDX2-elevated DNM3 inhibits the proliferation and metastasis of colon cancer cells via AKT signaling pathway. Cancer Manag Res. 2019;11:5799–5811. doi:10.2147/CMAR.S193805
5. Azvolinsky A. Colorectal cancer: to stack or sequence therapy? J Natl Cancer Inst. 2015;107(5):djv138–djv138. doi:10.1093/jnci/djv138
6. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics. 2016;17(1):47–62. doi:10.1038/nrg.2015.10
7. Melissari M-T, Grote P. Roles for long non-coding RNAs in physiology and disease. Pflügers Archiv Eur J Physiol. 2016;468(6):945–958. doi:10.1007/s00424-016-1804-y
8. Bach D-H, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152–166. doi:10.1016/j.canlet.2018.01.053
9. Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:93–95. doi:10.1016/j.steroids.2017.12.012
10. Liang WH, Li N, Yuan ZQ, Qian XL, Wang ZH. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol Carcinog. 2019;58(4):461–473. doi:10.1002/mc.22941
11. Liao J, Xie N. Long noncoding RNA DSCAM-AS1 functions as an oncogene in non-small cell lung cancer by targeting BCL11A. Eur Rev Med Pharm Sci. 2019;23(3):1087–1092. doi:10.26355/eurrev_201902_16998
12. Huang YL, Xu Q, Wang X. Long noncoding RNA DSCAM-AS1 is associated with poor clinical prognosis and contributes to melanoma development by sponging miR-136. Eur Rev Med Pharmacol Sci. 2019;23(7):2888–2897. doi:10.26355/eurrev_201904_17567
13. Ji D, Hu G, Zhang X, Yu T, Yang J. Long non-coding RNA DSCAM-AS1 accelerates the progression of hepatocellular carcinoma via sponging miR-338-3p. Am J Transl Res. 2019;11(7):4290–4302.
14. Liu X, Zhou Y, Ning YE, Gu H, Tong Y, Wang N. MiR-195-5p inhibits malignant progression of cervical cancer by targeting YAP1. OncoTargets Ther. 2020;13:931–944. doi:10.2147/OTT.S227826
15. Song S, Fajol A, Tu X, Ren B, Shi S. miR-204 suppresses the development and progression of human glioblastoma by targeting ATF2. Oncotarget. 2016;7(43):70058–70065. doi:10.18632/oncotarget.11732
16. Chu Y, Jiang M, Du F, et al. miR-204-5p suppresses hepatocellular cancer proliferation by regulating homeoprotein SIX1 expression. FEBS Open Bio. 2018;8(2):189–200. doi:10.1002/2211-5463.12363
17. Li Y, Chen R, Li Z, et al. miR-204 negatively regulates cell growth and metastasis by targeting ROBO4 in human bladder cancer. OncoTargets Ther. 2019;12:8515–8524. doi:10.2147/OTT.S205023
18. Hu WB, Wang L, Huang XR, Li F. MicroRNA-204 targets SOX4 to inhibit metastasis of lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2019;23(4):1553–1562. doi:10.26355/eurrev_201902_17114
19. Yin -J-J, Liang B, Zhan X-R. MicroRNA-204 inhibits cell proliferation in T-cell acute lymphoblastic leukemia by down-regulating SOX4. Int J Clin Exp Pathol. 2015;8(8):9189–9195.
20. Wu D, Pan H, Zhou Y, et al. Upregulation of microRNA-204 inhibits cell proliferation, migration and invasion in human renal cell carcinoma cells by downregulating SOX4. Mol Med Rep. 2015;12(5):7059–7064. doi:10.3892/mmr.2015.4259
21. Lin CM, Fang CL, Hseu YC, et al. Clinical and Prognostic Implications of Transcription Factor SOX4 in Patients with Colon Cancer. PLoS One. 2013;8(6):e67128. doi:10.1371/journal.pone.0067128
22. Liu J, Du W. LncRNA FENDRR attenuates colon cancer progression by repression of SOX4 protein. OncoTargets Ther. 2019;12:4287–4295. doi:10.2147/OTT.S195853
23. Hao XZ, Yang K. LncRNA MAGI2-AS3 suppresses the proliferation and invasion of non-small cell lung carcinoma through miRNA-23a-3p/PTEN axis. Eur Rev Med Pharmacol Sci. 2019;23(17):7399–7407. doi:10.26355/eurrev_201909_18848
24. Wu X, Xiao Y, Zhou Y, Zhou Z, Yan W. lncRNA SNHG20 promotes prostate cancer migration and invasion via targeting the miR-6516-5p/SCGB2A1 axis. Am J Transl Res. 2019;11(8):5162–5169.
25. Cui M, Chen M, Shen Z, Wang R, Fang X, Song B. LncRNA-UCA1 modulates progression of colon cancer through regulating the miR-28-5p/HOXB3 axis. J Cell Biochem. 2019;120(5):6926–6936. doi:10.1002/jcb.27630
26. Peng CL, Zhao XJ, Wei CC, Wu JW. LncRNA HOTAIR promotes colon cancer development by down-regulating miRNA-34a. Eur Rev Med Pharmacol Sci. 2019;23(13):5752–5761. doi:10.26355/eurrev_201907_18312
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.