Back to Journals » Cancer Management and Research » Volume 12
LncRNA DANCR-miR-758-3p-PAX6 Molecular Network Regulates Apoptosis and Autophagy of Breast Cancer Cells
Authors Zhang XH, Li BF, Ding J, Shi L, Ren HM, Liu K, Huang CC, Ma FX, Wu XY
Received 15 March 2020
Accepted for publication 6 May 2020
Published 29 May 2020 Volume 2020:12 Pages 4073—4084
DOI https://doi.org/10.2147/CMAR.S254069
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Xian Hu Zhang,1 Bing Feng Li,1 Jie Ding,2 Lei Shi,1 Huo Ming Ren,1 Kui Liu,1 Chuan Cai Huang,1 Fu Xiao Ma,1 Xin Yao Wu1
1Department of General Surgery, Suzhou First People’s Hospital, Suzhou, Anhui 234000, People’s Republic of China; 2Physical Examination Department, Suzhou Central Blood Station, Suzhou, Anhui 234000, People’s Republic of China
Correspondence: Xian Hu Zhang
Department of General Surgery, Suzhou First People’s Hospital, No. 26 Yinhe Road, Suzhou, Anhui 234000, People’s Republic of China
Tel +86 18134605661
Email [email protected]
Objective: This study set out to probe into the effects of long non-coding RNA (LncRNA) differentiation antagonizing non-protein coding RNA (DANCR) on apoptosis and autophagy of breast cancer (BC) cells.
Methods: The expression levels of DANCR, miR-758-3p and paired box 6 (PAX6) in BC tissues and cell lines were detected. The transcription and protein levels of PAX6, apoptosis-related factors (caspase-3, caspase-9, Bax/Bcl-2), and autophagy-related factors (LC3B, Atg5, Beclin-1) in BC cells were detected. The cell proliferation, apoptosis, autophagy and the regulatory relationship between genes and target genes were analyzed.
Results: DANCR and PAX6 were up-regulated in BC tissues and cell lines, while miR-758-3p was opposite. Down-regulating DANCR inhibited the malignant proliferation of BC cells and also promoted apoptosis and autophagy, which showed that caspase-3, caspase-9, Bax/Bcl-2, LC3B, Atg5 transcription and protein levels increased, while Beclin-1 transcription and protein levels decreased. DANCR regulated miR-758-3p in a targeted manner, and its over-expression could weaken the anti-cancer effect of miR-758-3p on BC cells. In addition, miR-758-3p also directly targeted PAX6, and knocking down its expression could weaken the inhibitory effect of down-regulating PAK6 on BC cell apoptosis and autophagy. We also found that DANCR acted as a competitive endogenous RNA sponge miR-758-3p, thus regulating the PAX6 expression.
Conclusion: DANCR-miR-758-3p-PAX6 molecular network plays a key regulatory role in BC cell apoptosis and autophagy, which may provide reference for treating patients.
Keywords: DANCR, miR-758-3p, PAX6, breast cancer, BC, apoptosis and autophagy
Introduction
Breast cancer (BC) is the main cause of female cancer death. The annual morbidity has an upward trend, and its occurrence factors include hormones, reproduction, and living habits.1,2 According to the epidemic data of BC, its lifetime risk is 12.30%, with 1,671,149 global cases and 521,907 deaths.3,4 At present, the prognosis of BC mainly depends on the stage of tumor progression and molecular subtypes. Early diagnosis is helpful to improve the cure rate or survival rate. However, there may be over-diagnosis and treatment problems. Therefore, it is still important and necessary to explore the pathological mechanism of BC.5 Research on BC-related molecular networks will help us to reveal the regulatory mechanism of transcription networks on tumor biological processes and find specific targeted therapeutic schemes.6,7 We will explore the development mechanism of BC from a molecular perspective, which has potential value for the prevention and treatment of patients.
The anti-cancer characteristics of anticancer drugs are often reflected in the suppression of tumor cells, including the inhibition of proliferation and the induction of apoptosis and autophagy. Apoptosis and autophagy belong to the anti-proliferation events of cells, and studying the two helps us understand the survival of tumor cells.8,9 Long non-coding RNA (LncRNA) is a long-chain RNA molecule that can change gene expression and can form LncRNA–microRNA molecular network to regulate cell events such as apoptosis and autophagy.10 Differentiation antagonist non-protein coding RNA (DANCR) belongs to LncRNA family, has carcinogenic activity in various tumors, and also acts as a molecular sponge of miR-216a-5p in BC to promote tumor progression.11,12 We found potential targeting sites between DANCR and miR-758-3p in StarBase, and also found that the molecular network of DANCR-miR-758-3p participated in the pathological mechanism of non-small cell lung cancer.13 miR-758-3p mediates the molecular network of CASC9-miR-758-3p-LIN7A, which has anti-cancer properties in ovarian cancer, but the regulatory mechanism in BC has not been elaborated in detail.14 By querying Targetscan7.2, we discovered that there were potential binding sites between paired box 6 (PAX6) and miR-758-3p. PAX6 belongs to the matched box transcription factor family, which can affect the behavior of tumor cells. As a carcinogenic factor in BC, PAX6 can promote the transfer of BC cells, but its regulatory mechanism on apoptosis and autophagy is still unclear.15,16
We suspect that the DANCR-miR-758-3p-PAX6 molecular network has regulatory effects on apoptosis and autophagy of BC cells, and hereby conduct research and report.
Materials and Methods
Collection of Tissue Samples
Forty-six BC patients admitted to Suzhou First People’s Hospital from February 2014 to February 2017 were selected. With the consent of the patients, cancerous tissues (BC group) and paracancerous tissues (normal group) were obtained during the operation and stored in liquid nitrogen tanks for standby. They were (42.34±6.25) years old on average. This test has been approved by the Ethics Committee of Suzhou First People’s Hospital. All the related parties agreed to participate in the test and sign an informed consent form. Inclusion criteria were as follows: those confirmed by pathology as BC;17 first treatment; not taking any drugs that affect the results of this study within half a year; people with normal cognition and no communication problems. Exclusion criteria were as follows: patients with other breast diseases; patients with other malignances; there were serious heart, lung, kidney and other organ complex diseases; patients with serious infectious diseases; patients who refused to provide experimental specimens.
Follow-Up
The patients were followed up for 3 years, mainly by telephones, visits, medical record inquiries, etc. And the overall survival (OS) was from treatment to death or the last day of follow-up.
Cell Culture and Transfection
We purchased human BC cell lines HCC1937, 1590, ZR-75-30, MDA-MB-468 and normal breast epithelial cells MCF-10A (Yubo Biotechnology Co., Ltd., Shanghai, China, YBCC102145, YBCC100935, YBCC100125, YBCC100687, AE-364). They were cultured in DMEM medium (Beiyuanxin Biotechnology Co., Ltd., Changzhou, China, BYX1540P) containing 10%PBS, 100 units/mL penicillin, and 100 μg/mL streptomycin (Cellmax Biotechnology Co., Ltd., Beijing, China, CPS101.02) at 37°C, 5%CO2.
By constructing recombinant plasmids, pEGFP-DANCR and pSilencer-DANCR/PAX6, we found that the expression levels were up-regulated and down-regulated, respectively. DANCR low expression plasmid pSilencer-DANCR (si-DANCR), DANCR high expression plasmid pEGFP-DANCR (DANCR), empty vector (vector), PAX6 low expression plasmid pSilencer-PAX6 (si-PAX6), negative control RNA (si-NC), miR-758-3p mimetic (miR-758-3p), miR negative control (miR-NC), anti-miR-758-3p sequence (anti-miR-758-3p), and anti-miR negative control (anti-miR-NC) were transfected into cells with Lipofectamine™ 2000 kit (BioMag Biotechnology Co., Ltd., Wuxi, China, 11668019), and the operation steps were strictly conducted based on the kit instructions.
Real-Time Quantitative PCR
The total RNA was extracted from tissues and cells by Trizol reagent (Chundu Biotechnology Co., Ltd., Wuhan, China, CD-102523GM). Each 5 μg of total RNA was taken to conduct reverse transcription cDNA operation in the light of the instructions of reverse transcription kit (Chundu Biotechnology Co., Ltd., Wuhan, China, CD-102539GM). After transcription, 1 μL of synthesized cDNA was taken for amplification. DANCR amplification system was as follows: cDNA 1 μL, upstream and downstream primers 0.4 μL each, 2X TransScript® Tip Green qPCR SuperMix 10 μL, Passive Reference Dye (50X) 0.4 μL, Nuclease-free Water supplemented to 20 μL. miR-758-3p amplification system was as follows: cDNA 1 μL, upstream and downstream primers 0.4 μL each, 2×TransTaq® Tip Green qPCR SuperMix 10 μL, Passive Reference Dye (50X) 0.4 μL, ddH2O supplemented to 20 μL. PCR reaction conditions were as follows: pre-denaturation at 94°C for 30 s, denaturation at 94°C for 5 s, annealing extension at 60°C for 30 s, 40 cycles in total. mRNA employed β-Actin as internal reference, miRNA employed U6 as internal reference, and data were analyzed by 2−ΔΔct.
Western Blot Test
The total protein of tissues and cultured BC cells was extracted by RIPA lysis method, and the protein concentration was detected by BCA kit (Boppard Trading Co., Ltd., Guangzhou, China, 297–73101). After that, the protein concentration was adjusted to 4 μg/μL, the protein was separated by electrophoresis and transferred to PVDF membrane, closed with 5% skimmed milk, and then incubated with primary antibody at 4°Call night. The dilution ratio of PAX6, caspase-3, caspase-9, Bax/Bcl-2, LC3B, Atg5, Beclin-1, β-Actin and other primary antibodies was 1:1000. We used rabbit polyclonal antibody to detect all primary antibodies. The antibodies were purchased from Otwo Biotech (Shenzhen) Inc. Each membrane was rinsed 3 times with PBS (Qiangxin Biorepublic Co., Ltd., Beijing, China, BSS-1005-A) for 15 min each time, and then it was incubated with horseradish peroxidase-labeled goat anti-rabbit secondary antibody. Excess liquid on the membrane was absorbed by filter paper. Afterwards, it developed in a darkroom using the enhanced chemiluminescence (ECL). Finally, we analyzed the gray value.
Cell Viability Detected by MTT Assay
The cells transfected for 24 h were inoculated on 96-well plates, and the cell density was adjusted to 4×103 cells/well. They were incubated 0, 24, 48 and 72 h, respectively, at 37°C, and then we supplemented MTT solution 20 μL (5 μmg/mL) (Baiaolaibo Technology Co., Ltd., Beijing, China, GL0247-SBJ). Afterwards, they were further cultured 4 h at 37°C, and then we supplemented dimethyl sulfoxide (200 μL) to each well. The OD value of cells in each group was measured at a wavelength of 450 mm using a Photopette spectrophotometer (Pubiao Experimental Equipment Technology Co., Ltd., Dongguan, China, SPCC).
Apoptosis Detected by Flow Cytometry
Cells were made into 1×106/mL suspension after being digested with 0.25% trypsin, cleaned twice with PBS, mixed with 100 μL binding buffer, etc. After that, 10 μL of Annex5minV-FITC and 10 μL of PI were supplemented to the suspension. The suspension was incubated 5 min at room temperature in the dark and detected by NovBCyte flow cytometry (Derica Biotechnology Co., Ltd., Beijing, China, DLK0002051). The test was repeated 3 times and results were averaged.
Dual-Luciferase Report Assay
Complementary DNA fragments of DANCR wild type (DANCR-Wt) and DANCR mutant (DANCR-Mut) fragments, miR-758-3p-Wt and miR-758-3p-Mut fragments were subcloned downstream of the luciferase gene in the luciferase report vector, respectively. miR-758-3p was co-transfected with PAX6-Wt or PAX6-Mut. After 2d, fireflies and renin luciferase activities in cell lysates were continuously measured via a dual-luciferase report kit (Chundu Biotechnology Co., Ltd., Wuhan, China, CDLG-4997).
RNA Immunoprecipitation
RNA immunoprecipitation (RIP) was employed to verify the interaction between DANCR and miR-758-3p. The experiment was carried out via the instructions of EZMagna RIP kit (Guyan Biotech Co., Ltd., Shanghai, China, GOY-E5944). Cells were lysed, incubated with protein A magnetic beads, and conjugated with antibodies at 4°C. After 6 h, the beads were washed and then incubated 30 min with 0.1%SDS/0.5 mg/mL protease K at 55°C. At last, the immunoprecipitated RNA was analyzed via specific primers to prove the existence of DANCR and miR-758-3p.
RNA Pull-Down Experiment
BC cells were transfected with biotinylated miR-758-3p-Wt, miR-758-3p-Mut and negative control Bio-NC, respectively. After 2d, the cell lysate was incubated with Streptomyces M-280 magnetic beads based on the production instructions, and then the DANCR level in the RNA complex bound to the beads was detected.
Statistical Methods
In this study, GraphPad 6 software package was used for data analysis and picture drawing. Independent-samples t-test, one-way analysis of variance (ANOVA), LSD-t test, repeated measures ANOVA and Bonferroni were used for comparison between the two groups, comparison among multiple groups, post hoc pairwise comparison, comparison of multi-time indexes, backtesting, respectively. Correlation was analyzed by Pearson test, and the independent risk factors influencing the prognosis of patients were analyzed via Multivariate Cox regression. Survival curve was drawn via Kaplan–Meier method, and the difference of survival time between groups was evaluated via Log rank test. P<0.05 means statistical difference.
Results
DANCR Is Up-Regulated in BC Samples and Cell Lines
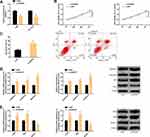
DANCR was up-regulated in cancer tissue samples of BC patients and those with poor prognosis. All the 46 patients completed the follow-up successfully, and the 3-year survival rate was 63.04% (29/46). The high expression of DANCR was remarkably correlated with the lower 3-year OS of BC patients. In addition, we also observed up-regulation of DANCR in BC cell line, and it was more obvious in 1590 and ZR-75-30. We will further analyze these two cells. The above results have statistical significance (P<0.05). (Figure 1)
Knocking Down DANCR Is Not Conducive to BC Cell Proliferation, but Induces Apoptosis and Autophagy
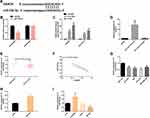
When si-DANCR was transfected into BC cells, DANCR was markedly down-regulated, cell proliferation behavior was suppressed, apoptosis level increased, caspase-3, caspase-9, Bax/Bcl-2, LC3B, Atg5 transcription and protein level all increased, while Beclin-1 transcription and protein level all decreased, and the difference was statistically remarkable (P<0.05). (Figure 2)
DANCR Has Targeted Relationship with miR-758-3p
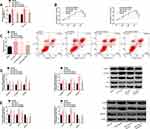
We discovered that DANCR and miR-758-3p had potential target sites through starBase. After miR-758-3p was up-regulated, only DANCR-Wt luciferase activity (instead of DANCR-Mut) decreased obviously. In RIP experiment, the DANCR and miR-758-3p levels precipitated by Ago2 antibody were markedly higher than IgG. In RNA pull-down experiment, DANCR was only pulled down by biotin-labeled miR-758-3p-WT. Soon afterwards, clinically, the cancer tissues of BC patients have significantly lower levels of miR-758-3p, and DANCR is markedly negatively correlated with miR-758-3p (r=−0.723, P<0.001). In BC cell line, we found that the miR-758-3p level was also low and its level in 1590 and ZR-75-30 was lower, so we made further analysis with 1590 and ZR-75-30. In BC cells transfected with DANCR, the DANCR expression was enhanced, and the miR-758-3p expression was blocked while enhanced after si-DANCR was transfected. The above cell analysis has statistical significance (P<0.05). (Figure 3)
DANCR Reverses Antitumor Effect of miR-758-3p on BC Cells
The miR-758-3p expression was enhanced after it was transfected into BC cells, while it was weakened after transfection of DANCR. We also found that the proliferation of BC cells was inhibited and apoptosis was induced after transfection of miR-758-3p mimics; caspase-3, caspase-9, Bax/Bcl-2, LC3B, Atg5 transcription and protein level all increased, while Beclin-1 transcription and protein level all decreased. However, the influence of miR-758-3p on BC cells reduced after further transfection of DANCR. The above results have statistical significance (P<0.05). (Figure 4)
miR-758-3p Targets PAX6
There were potential binding sites between miR-758-3p and PAX6, and up-regulating miR-758-3p only reduced PAX6-Wt luciferase activity obviously. The expression and protein level of PAX6 in BC cancer tissues were remarkably higher than those in paracancerous tissues and were markedly negatively correlated with miR-758-3p (r=−0.626, P<0.001). The expression and protein level of PAX6 in BC cells were also markedly higher, especially in 1590 and ZR-75-30, which were hereby used for further analysis. The miR-758-3p expression in cells was inhibited after transfection of anti-miR-758-3p, but the expression levels of PAX6 and protein level were improved, while the transfection of miR-758-3p mimics revealed the converse results. The above analysis has statistical significance (P<0.05). (Figure 5)
Down-Regulating miR-758-3p Can Weaken the Effect of Si-PAX6 on BC Cell Proliferation, Apoptosis and Autophagy
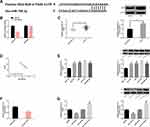
Mediated by si-PAX6, mRNA and protein levels of PAX6 reduced, BC cell proliferation was inhibited, apoptosis was promoted, and caspase-3, caspase-9, Bax/Bcl-2, LC3B, Atg5 transcription and protein level increased, while Beclin-1 transcription and protein level decreased. When miR-758-3p was further down-regulated, the above influence was weakened. All results were statistically remarkable (P<0.05). (Figure 6)
PAX6 Expression Is Regulated by DANCR and miR-758-3p
After transfection of miR-758-3p, PAX6 transcription and protein level were inhibited, while after further transfection of DANCR, PAX6 transcription and protein level were improved, and the difference was statistically remarkable (P<0.05). (Figure 7)
Discussion
BC is a cancer with highly specific clinical symptoms. It has a high morbidity in developed countries and a high mortality in developing countries.4,18 LncRNA is relevant to tumor progression, hormone change and chemical drug resistance of BC. Studying LncRNA is helpful for us to deeply understand the pathological development of BC.19 This time, we focus on the potential molecular regulatory network of LncRNA-DANCR in BC, hoping to provide valuable insights for treating patients.
More and more studies show that DANCR has carcinogenicity in BC. For instance, Zhang et al20 pointed out that DANCR-SOCS3-EZH2 molecular network could regulate the inflammatory phenotype and metastasis of BC cells, and up-regulating SOCS3 or down-regulating EZH2 is helpful to inhibit the carcinogenic activity of DANCR in BC cells. And Sha et al21 pointed out that knocking down the DANCR expression is conducive to inhibit the malignant progression of triple-negative BC. In this study, we evaluated the DANCR expression in BC patients’ tissues and cells. The results manifested that DANCR in BC cancer tissues and cell lines showed a high level, indicating that it might be effective in the disease progression of BC. Besides, we found that patients with poor prognosis had higher levels of DANCR. Our survival analysis also signified that the 3-year survival rate of BC patients was 63.04%, and the high level of DANCR was significantly related to the lower 3-year OS, suggesting that it was tied to the lower survival rate. Yoo et al22 studied that the 3-year survival rate of BC patients was about 62.60%, which was similar to our results. The focus of this study is apoptosis and autophagy of BC cells, both of which are induction methods of cell death. Increasing the level of apoptosis and autophagy of BC cells is helpful to reverse tumor progression.23,24
Caspase-3, caspase-9 and Bax have pro-apoptotic activity, while Bcl-2 has anti-apoptotic activity. High levels of caspase-3, caspase-9 and Bax/Bcl-2 indicate that cells are in a state of induced apoptosis.25,26 It is reported that caspase-3 and Bax can respond to ganoderma lucidum spore oil in vitro and in vivo treatment of BC. The inactivated caspase-9 is related to the tumor survival of triple-negative BC, and Bcl-2 is a vital factor in the anti-apoptosis defense mechanism of BC.27–29 Similarly, LC3B and Atg5 are positive regulators of autophagy, while Beclin-1 is negative regulator of autophagy. Among them, LC3B is tied to BC immune infiltration, Atg5 is tied to BC tumor occurrence and metastasis, and Beclin-1 is tied to BC patient survival and chemotherapy sensitivity.30–32 In this study, the above apoptosis and autophagy-related factors were measured to study the death events of BC cells. In the cell function test of DANCR, down-regulating DANCR is conductive to inhibit the proliferation of BC cells, induce apoptosis and autophagy, suggesting that its down-regulation is beneficial to inhibit the malignant progression of BC. In the molecular network of LncRNA-mRNA, the mechanism of action between LncRNA and mRNA is often that LNC RNA acts as a molecular sponge of mRNA to negatively regulate its gene expression, thus mediating the pathophysiological mechanism of the body.33 We have verified whether DANCR is a molecular sponge of miRNA. Dual-luciferase report, RIP and RNA pull-down experiments have all confirmed that DANCR and miR-758-3p have exact target-controlled relationship, and we have detected that miR-758-3p is highly expressed in BC tissues and cells. In correlation analysis, DANCR also has marked negative correlation with miR-758-3p. Further exploration of miR-758-3p’s effect on BC cell behavior found that its overexpression significantly inhibited the proliferation of BC cells and enhanced their apoptosis and autophagy, suggesting that it had anti-tumor activity in BC.
We further explored the regulatory mechanism of miR-758-3p in BC progression, predicted potential targets through TargetScan, and found potential binding sites between PAX6 and miR-758-3p. We further verified the binding relationship between them through the dual-luciferase report. PAX6 was associated with tissue proliferation and was frequently expressed abnormally in many tumors such as BC, which was equivalent to nutrients for cancer cell survival.34 Studies have reported that PAX6 is targeted by miR-375, and inhibiting its expression is beneficial to inhibit BC tumor metastasis.35 Our research found that inhibiting PAX6 expression was not conducive to BC cell proliferation, but was instrumental in apoptosis and autophagy. When we further down-regulated miR-758-3p, the above results had been significantly reversed, suggesting that PAX6 had regulatory effect on BC cell behavior, but this effect was also affected by miR-758-3p. Finally, we also found that the PAX6 expression was positively regulated by DANCR and negatively regulated by miR-758-3p.
Although this study confirmed that DANCR, as endogenous RNA, positively regulated PAX6 expression through competitive binding of miR-758-3p. Nevertheless, there is still room for improvement. First of all, we can analyze whether DANCR is an independent prognostic factor for BC and further explore whether it has the potential to predict its prognosis. What is more, we can also analyze whether DANCR has influence on BC chemical resistance and clarify its potential regulatory mechanism on BC chemical sensitivity. Furthermore, we can supplement the potential value of DANCR in predicting BC recurrence.
Conclusion
We first propose that there is a DANCR-miR-758-3p-PAX6 molecular regulatory network that mediates the biological function of BC cells, which may be of directive significance to the research and development of anti-tumor drugs for patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Mariotto AB, Etzioni R, Hurlbert M, Penberthy L, Mayer M. Estimation of the number of women living with metastatic breast cancer in the United States. Cancer Epidemiol Biomarkers Prev. 2017;26(6):809–815. doi:10.1158/1055-9965.EPI-16-0889
2. Iacoviello L, Bonaccio M, de Gaetano G, Donati MB. Epidemiology of breast cancer, a paradigm of the “common soil” hypothesis. Semin Cancer Biol. 2020. doi:10.1016/j.semcancer.2020.02.010
3. Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac J Cancer Prev. 2016;17(S3):43–46. doi:10.7314/APJCP.2016.17.S3.43
4. Rojas K, Stuckey A. Breast cancer epidemiology and risk factors. Clin Obstet Gynecol. 2016;59(4):651–672. doi:10.1097/GRF.0000000000000239
5. Winters S, Martin C, Murphy D, Shokar NK. Breast cancer epidemiology, prevention, and screening. Prog Mol Biol Transl Sci. 2017;151:1–32.
6. Alcala-Corona SA, de Anda-jauregui G, Espinal-Enriquez J, Hernandez-Lemus E. Network modularity in breast cancer molecular subtypes. Front Physiol. 2017;8:915. doi:10.3389/fphys.2017.00915
7. Chereda H, Bleckmann A, Kramer F, Leha A, Beissbarth T. Utilizing molecular network information via graph convolutional neural networks to predict metastatic event in breast cancer. Stud Health Technol Inform. 2019;267:181–186. doi:10.3233/SHTI190824
8. Sung B, Chung HY, Kim ND. Role of apigenin in cancer prevention via the induction of apoptosis and autophagy. J Cancer Prev. 2016;21(4):216–226. doi:10.15430/JCP.2016.21.4.216
9. Yu P, Zhang C, Gao CY, et al. Anti-proliferation of triple-negative breast cancer cells with physagulide P: ROS/JNK signaling pathway induces apoptosis and autophagic cell death. Oncotarget. 2017;8(38):64032–64049. doi:10.18632/oncotarget.19299
10. Lv Y, Liu Z, Huang J, Yu J, Dong Y, Wang J. LncRNA nuclear-enriched abundant transcript 1 regulates hypoxia-evoked apoptosis and autophagy via mediation of microRNA-181b. Mol Cell Biochem. 2020;464(1–2):193–203. doi:10.1007/s11010-019-03660-2
11. Thin KZ, Liu X, Feng X, Raveendran S, Tu JC. LncRNA-DANCR: a valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract. 2018;214(6):801–805. doi:10.1016/j.prp.2018.04.003
12. Tao W, Wang C, Zhu B, Zhang G, Pang D. LncRNA DANCR contributes to tumor progression via targetting miR-216a-5p in breast cancer: lncRNA DANCR contributes to tumor progression. Biosci Rep. 2019;39(4). doi:10.1042/BSR20181618
13. Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. 2018;103:94–100. doi:10.1016/j.biopha.2018.03.053
14. Hu X, Li Y, Kong D, Hu L, Liu D, Wu J. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J Cell Physiol. 2019;234(7):10800–10808. doi:10.1002/jcp.27903
15. Wang J, Jia N, Lyv T, et al. Paired box 2 promotes progression of endometrial cancer via regulating cell cycle pathway. J Cancer. 2018;9(20):3743–3754. doi:10.7150/jca.22418
16. Urrutia G, Laurito S, Campoy E, Nasif D, Branham MT, Roque M. PAX6 promoter methylation correlates with MDA-MB-231 cell migration, and expression of MMP2 and MMP9. Asian Pac J Cancer Prev. 2018;19(10):2859–2866. doi:10.22034/APJCP.2018.19.10.2859
17. Cardoso F, Kyriakides S, Ohno S, et al. Early breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30(8):1194–1220. doi:10.1093/annonc/mdz173
18. Dai X, Cheng H, Bai Z, Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. 2017;8(16):3131–3141. doi:10.7150/jca.18457
19. Niknafs YS, Han S, Ma T, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7(1):12791. doi:10.1038/ncomms12791
20. Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. 2020;14(2):309–328. doi:10.1002/1878-0261.12622
21. Sha S, Yuan D, Liu Y, Han B, Zhong N. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol Open. 2017;6(9):1310–1316. doi:10.1242/bio.023135
22. Yoo TK, Chae BJ, Kim SJ, et al. Identifying long-term survivors among metastatic breast cancer patients undergoing primary tumor surgery. Breast Cancer Res Treat. 2017;165(1):109–118. doi:10.1007/s10549-017-4309-2
23. Chen Q, Kang J, Fu C. The independence of and associations among apoptosis, autophagy, and necrosis. Signal Transduct Target Ther. 2018;3(1):18. doi:10.1038/s41392-018-0018-5
24. Sun ZL, Dong JL, Wu J. Juglanin induces apoptosis and autophagy in human breast cancer progression via ROS/JNK promotion. Biomed Pharmacother. 2017;85:303–312. doi:10.1016/j.biopha.2016.11.030
25. Sirpu Natesh N, Arumugam M, Karanam G. Apoptotic role of marine sponge symbiont Bacillus subtilis NMK17 through the activation of caspase-3 in human breast cancer cell line. Mol Biol Rep. 2018;45(6):2641–2651. doi:10.1007/s11033-018-4434-y
26. Do Nascimento-Neto LG, Cabral MG, Carneiro RF, et al. Halilectin-3, a lectin from the marine sponge haliclona caerulea, induces apoptosis and autophagy in human breast cancer MCF7 cells through caspase-9 pathway and LC3-II protein expression. Anticancer Agents Med Chem. 2018;18(4):521–528. doi:10.2174/1871520617666171114094847
27. Jiao C, Chen W, Tan X, et al. Ganoderma lucidum spore oil induces apoptosis of breast cancer cells in vitro and in vivo by activating caspase-3 and caspase-9. J Ethnopharmacol. 2020;247:112256. doi:10.1016/j.jep.2019.112256
28. Yang J, Meng X, Yu Y, Pan L, Zheng Q, Lin W. LncRNA POU3F3 promotes proliferation and inhibits apoptosis of cancer cells in triple-negative breast cancer by inactivating caspase 9. Biosci Biotechnol Biochem. 2019;83(6):1117–1123. doi:10.1080/09168451.2019.1588097
29. Konig SM, Rissler V, Terkelsen T, Lambrughi M, Papaleo E. Alterations of the interactome of Bcl-2 proteins in breast cancer at the transcriptional, mutational and structural level. PLoS Comput Biol. 2019;15(12):e1007485. doi:10.1371/journal.pcbi.1007485
30. Ladoire S, Enot D, Senovilla L, et al. The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy. 2016;12(5):864–875. doi:10.1080/15548627.2016.1154244
31. Han Q, Deng Y, Chen S, et al. Downregulation of ATG5-dependent macroautophagy by chaperone-mediated autophagy promotes breast cancer cell metastasis. Sci Rep. 2017;7(1):4759. doi:10.1038/s41598-017-04994-x
32. Gu Y, Chen T, Li G, et al. Lower Beclin 1 downregulates HER2 expression to enhance tamoxifen sensitivity and predicts a favorable outcome for ER positive breast cancer. Oncotarget. 2017;8(32):52156–52177. doi:10.18632/oncotarget.11044
33. Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148–64167. doi:10.18632/oncotarget.11637
34. Muratovska A, Zhou C, He S, Goodyer P, Eccles MR. Paired-Box genes are frequently expressed in cancer and often required for cancer cell survival. Oncogene. 2003;22(39):7989–7997. doi:10.1038/sj.onc.1206766
35. Zou Q, Yi W, Huang J, Fu F, Chen G, Zhong D. MicroRNA-375 targets PAX6 and inhibits the viability, migration and invasion of human breast cancer MCF-7 cells. Exp Ther Med. 2017;14(2):1198–1204. doi:10.3892/etm.2017.4593
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.