Back to Journals » Cancer Management and Research » Volume 12
LncRNA CDKN2B-AS1 Promotes Cell Viability, Migration, and Invasion of Hepatocellular Carcinoma via Sponging miR-424-5p
Authors Shen X, Li Y, He F, Kong J
Received 25 November 2019
Accepted for publication 6 June 2020
Published 4 August 2020 Volume 2020:12 Pages 6807—6819
DOI https://doi.org/10.2147/CMAR.S240000
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lu-Zhe Sun
Xinying Shen, Yong Li, Fan He, Jian Kong
Department of Interventional Radiology, Shenzhen People’s Hospital, Shenzhen, Guangdong, People’s Republic of China
Correspondence: Xinying Shen
Department of Interventional Radiology, Shenzhen People’s Hospital, Luohu District, Shenzhen, Guangdong 518020, People’s Republic of China
Tel +86 755-22948106
Email [email protected]
Objective: Hepatocellular carcinoma (HCC) results in high mortality and metastasis. In this study, the effects of long non-coding RNA (lncRNA) CDKN2B-AS1 on the progression of HCC were investigated.
Materials and Methods: LncRNA CDKN2B-AS1 expression of HCC cancer and adjacent tissues, and HCC cells were detected. Subsequently, CDKN2B-AS1 was overexpressed and silenced in HCC cells to observe the effects of CDKN2B-AS1 on the cell viability, migration, invasion, and epithelial–mesenchymal transition (EMT) of HCC cells by performing cell counting kit-8 (CCK-8), wound-healing, Transwell, and Western blot. The target gene of CDKN2B-AS1 was predicted and verified to be miR-424-5p, whose expression in HCC cells with up- or down-regulation of CDKN2B-AS1 expression was determined. Moreover, the effects of miR-424-5p on cell viability, migration, and invasion and EMT of HCC cells were investigated with miR-424-5p up-regulation or down-regulation, together with overexpression or silencing of CDKN2B-AS1.
Results: CDKN2B-AS1 expression was increased in HCC tissues and cells. Silencing of CDKN2B-AS1 suppressed cell viability, migration, invasion, and EMT, while overexpression of CDKN2B-AS1 produced the opposite results. Furthermore, CDKN2B-AS1 was predicted and verified to target miR-424-5p and was confirmed to negatively modulate miR-424-5p expression. Moreover, overexpression of miR-424-5p partially suppressed the previously high cell viability, migration, and invasion, and activated EMT resulted from up-regulation of CDKN2B-AS1, while silencing of miR-424-5p elevated the cellular processes inhibited by silencing the expression of CDKN2B-AS1.
Conclusion: The present study revealed that high-expressed CDKN2B-AS1 may associate with the progression of HCC by affecting the cell viability, migration, invasion, and EMT of HCC cells by negatively regulating miR-424-5p.
Keywords: lncRNA CDKN2B-AS1, miR-424-5p, migration, invasion, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) has high mortality and metastasis, and approximately 626 000 new cases of HCC were newly diagnosed annually.1 Surgery resection is the main method for treating early stage HCC. As HCC only presents few symptoms in its early stage, most HCC patients with more obvious symptoms are already at middle or advanced stage with tumor metastasis by the time of their diagnosis,2 and surgery resection as well as chemotherapies and radiotherapies at this time have limited effects on preventing postoperative recurrence and metastasis.3 Thus, studies on molecular mechanisms of its initiation and development of HCC and the new diagnostic target are highly necessary.
Long non-coding RNAs (lncRNAs) are RNA with approximately 200 nucleotides but do not code proteins. LncRNAs are increasingly studied for its potential involvement in cancers. LncRNAs play important roles in cancers through regulating the tumorigenesis, development, and prognosis of cancers.4,5 Previous studies reported that lncRNAs are dysregulated in different types of cancers and have critical effects related to cancer biology.6–9 Some lncRNAs have been reported to be closely related to liver cancer pathology, progression, prognosis, and the maintenance of cancer stem cell-like properties.10,11 For example, LINC00668 up-regulation accelerates cell proliferation, migration, and invasion of HCC via targeting miR-532-5p/YY1 axis;12 HAGLROS is high-expressed in HCC and its high expression is correlated with the progression of HCC by affecting cell proliferation, apoptosis, and autophagy via regulating miR-5095/ATG12 axis;13 HCC patients with high-expressed lncRNA PDZD7 tend to show a poorer prognosis; PDZD7 promotes stemness properties and inhibits chemosensitivity of HCC via regulating the miR-101/EZH2/ATOH8 pathway.14 lncRNAs closely associated with HCC should be further investigated for discovering novel promising biomarkers and potential targets for HCC treatment.
lncRNA CDKN2B antisense RNA 1 (CDKN2B-AS1), which is an antisense of the cyclin-dependent kinase inhibitor 2B (CDKN2B), plays important roles in various diseases including in cancers15–18. Zhu et al reported that interference of CDKN2B-AS1 restrains the metastasis and promotes apoptosis and senescence of cervical cancer cells via regulating miR-181a-5p/TGFβI axis.19 CDKN2B-AS1 facilitates osteosarcoma progression by sponging miR-4458 to increase MAP3K3 expression.17 However, the role of CDKN2B-AS1 in HCC has not been studied yet.
In this study, the role of CDKN2B-AS1 in relation to viability, migration and invasion, and EMT of HCC cells were investigated. Moreover, the relationship of CDKN2B-AS1 with low-expressed miR-424-5p in HCC was explored.
Materials and Methods
Cells and Tissues
Human hepatocyte cell line THLE-2 (CRL-2706) and HCC cell line (Huh7 (PTA-4583), Hep3B (HB-8064), and Sk-Hep1 (HTB-52)) were purchased from American Type Culture Collection (ATCC, USA), and MHCC97H (BNCC346681) was obtained from BeNa Culture Collection (Beijing). All the cell lines were cultivated in RPMI-1640 medium (Gibco, Rockville, MD) containing with 10% FBS (Thermo Scientific HyClone, Beijing, China) in a humidified incubator at 37°C.
Human normal HCC (n=30) and its adjacent tissues (n=30) were collected from HCC patients who did not receive the pretreatment of preoperative radiotherapy or chemotherapy before the surgery from 01/08/2016 to 30/06/2018 in Shenzhen People’s Hospital. Clinical and pathological characteristics were obtained from patient charts. The clinicopathological features are shown in Table 1. All the participants in this study allowed their tissues to be used for research and signed informed consent. The study (2016) was approved by the Ethics Committee of Shenzhen People’s Hospital.
 |
Table 1 The Correlation Between CDKN2B-AS1 Expression and Clinicopathological Characteristics of Patients with Hepatocellular Carcinoma |
CCK-8 Assay
The transfected Huh7 and MHCC97H cells were incubated in a 96-well culture plate at 5 103 cells/well. After incubation for 24 h, 48 h, and 72 h, the cell viability was determined by cell counting kit-8 (CCK-8) (Dojindo) according to the manufacture’s protocol. The absorbance was measured at 450 nm using Elx800 Reader (Bio-Tek Instruments Inc., VT, USA).
103 cells/well. After incubation for 24 h, 48 h, and 72 h, the cell viability was determined by cell counting kit-8 (CCK-8) (Dojindo) according to the manufacture’s protocol. The absorbance was measured at 450 nm using Elx800 Reader (Bio-Tek Instruments Inc., VT, USA).
Wound-Healing Assay
An amount of 5 105 cells was seeded into 6-well plates and pre-cultured for 24 h until its confluence reached at 90%. The surface was scratched using a sterile pipette tip to create a wound. After incubation for 48 h, migrated cells were counted under an inverted microscope (Olympus, Japan) and photographed.
105 cells was seeded into 6-well plates and pre-cultured for 24 h until its confluence reached at 90%. The surface was scratched using a sterile pipette tip to create a wound. After incubation for 48 h, migrated cells were counted under an inverted microscope (Olympus, Japan) and photographed.
Transwell Assay
An amount of 1 105 cells were seeded into Matrigel-coated upper chamber (8-μm pore size and a diameter of 6.5 mm) (Corning, USA), while the lower chamber was filled with medium containing with 10% FBS and stored under 37°C with 5% CO2 for 48 h. Next, the cells that did not invade through the pores were removed by a cotton swab. The cells on the bottom of the filter were fixed by 4% paraformaldehyde and dyed by crystal violet, and then counted under an inverted microscope.
105 cells were seeded into Matrigel-coated upper chamber (8-μm pore size and a diameter of 6.5 mm) (Corning, USA), while the lower chamber was filled with medium containing with 10% FBS and stored under 37°C with 5% CO2 for 48 h. Next, the cells that did not invade through the pores were removed by a cotton swab. The cells on the bottom of the filter were fixed by 4% paraformaldehyde and dyed by crystal violet, and then counted under an inverted microscope.
Cell Transfection
The full length of CDKN2B-AS1 sequence in the synthesized pcDNA3.1 vector and its negative control was obtained from Geneseed Biotech (Guangzhou, China). An equal amount of siCDKN2B-AS1 (siB13812144322-1-5) and its negative control, miR-424-5p mimic (miR10001341-1-5), miR-424-5p (miR20001341-1-5) inhibitor and their negative controls were purchased from Rib-Bio (China). The Huh7 and MHCC97H cells were pre-cultured and transfected using Lipofectamine TM 2000 transfection reagent (#11,668,019, ThermoFisher, USA) following the manufacturer’s instructions.
Real-Time Quantitative PCR (RT-qPCR)
Total RNAs of CDKN2B-AS1 were extracted from HCC and its adjacent tissues, transfected Huh7 and MHCC97H cells with TRIzol reagent (Invitrogen), and then reverse-transcribed into cDNAs by RevertAid First Strand cDNA Synthesis Kit (ThermoFisher Scientific, USA). The PCR process was performed in Step One Plus Real-Time PCR system (Applied Biosystems, Carlsbad, USA) under the following conditions: pre-denaturation at 95°C for 10 min, followed with 40 cycles at 95°C for 10 s, and annealing at 58°C for 35 s, extended at 72°C for 10 s. For determining miR-424-5p expression of transfected Huh7 and MHCC97H cells, and RT-qPCR analysis of miR-424-5p was prepared with an All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, Maryland, USA), according to the manufacturer’s instructions. GAPDH and U6 were used as internal references for CDKN2B-AS1 and miR-424-5p, respectively, and normalized by 2-ΔΔCT method. The primer sequences used are shown in Table 2.
 |
Table 2 The Primers Sequence Used for qPCR |
Western Blot
Cells were lysed on ice using RIPA buffer (Beyotime, Shanghai, China). The proteins were harvest and their concentration was determined by BCA Assay Kit (Pierce Chemical Co., Rockford, IL, USA). The proteins were separated by 10% SDS-PAGE and then transferred onto polyvinylidene fluoride membranes (PVDF, Millipore, Billerica, MA, USA), which were sealed by 5% skimmed milk for 1 h. Then, the membranes were incubated with the following primary antibodies at 4°C overnight: E-Cadherin (CST, #14,472, 1:1000, USA), N-Cadherin (CST, #14,215, 1:1000, USA), and Snail (Abcam, ab53519, 1:2000, UK). After washing, the membranes were incubated with secondary goat anti-rabbit (Abcam, ab205719, 1:1000, UK) and donkey anti-goat (Abcam, ab205723, 1:2000, UK) for 2 h at room temperature. After rinsing, the membranes were visualized by ECL reagent (Cell Signaling Technology, USA) and examined by Amersham Imager 600 (GE Healthcare Life Sciences). The data were analyzed by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Target Gene Prediction and Verification by Dual-Luciferase Reporter Assay
Target gene of CDKN2B-AS1 was predicted by starBase and then verified by dual-luciferase reporter assay. The wild type of 3ʹ-UTR of CDKN2B-AS1 was amplified and sub-cloned into pMIR-GLO luciferase vector (Promega, USA), and the contractive mutant 3ʹUTR fragment was constructed and inserted into pMIR-GLO luciferase vector. The WT or MUT 3ʹ-UTR of CDKN2B-AS1 were co-transfected into Huh7 and MHCC97H cells using Lipofectamine 2000 (ThermoFisher, USA). The luciferase activities of the transfected cells were measured by dual-luciferase reporter assay system (Promega, USA). Renilla luciferase activity was used for reference.
Statistical Analysis
The data were shown as means ± standard deviation and analyzed by SPSS. The Student’s t-test was used for the comparison between two groups, while one-way ANOVA was conducted for multiple comparisons. All the experiments were independently repeated in triplicate. P<0.05 was considered as statistically different.
Results
CDKN2B-AS1 Expression in HCC Tissues and Cells and Was Correlated with Clinicopathological Features
CDKN2B-AS1 expression in HCC and its adjacent tissues, human normal hepatocyte, and HCC cells was measured by RT-qPCR. The results showed that CDKN2B-AS1 expression in 30 HCC tissue samples was increased compared with that in the adjacent tissues (P<0.001, Figure 1A). As shown in Figure 1B, CDKN2B-AS1 expression was significantly high in HCC cell lines compared with that in normal hepatocyte THLE-2 (P<0.001). The result of associated association between the expression of CDKN2B-AS1 and clinical features of HCC patients showed that CDKN2B-AS1 was significantly correlated with clinical stage (P< 0.05, Table 1).
Effect of CDKN2B-AS1 on Viability, Migration, Invasion, and EMT of Huh7 and MHCC97H Cells
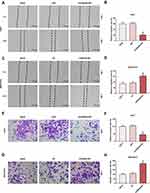
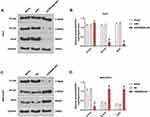
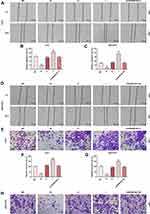
The siCDKN2B-AS1 and CDKN2B-AS1 were transfected into Huh7 cells and MHCC97H cells, respectively, and the transfection efficiency was detected. The data showed that the expression level of CDKN2B-AS1 was significantly low in Huh7 cells but significantly high in MHCC97H cells (Figure 2A and B). For the viabilities of Huh7 and MHCC97H cells (Figure 2C and D), and the results showed that the viability of Huh7 cells transfected with siCDKN2B-AS1 was greatly low, but that of MHCC97H cells transfected with CDKN2B-AS1 was significantly high (P<0.001). Cell migration and invasion experiment results revealed that the migration and invasion of Huh7 cells with silencing CDKN2B-AS1 were reduced, but while those of MHCC97H with overexpressed CDKN2B-AS1 were significantly increased (Figure 3). Meanwhile, the expressions of proteins related to EMT in Huh7 and MHCC97H cells were determined. We found that silencing expression of CDKN2B-AS1 reduced N-Cadherin and Snail expressions and increased E-Cadherin expression in Huh7 cells (Figure 4A and B), overexpression of CDKN2B-AS1 increased the expressions of N-Cadherin and Snail and reduced E-Cadherin expression in MHCC97H cells (Figure 4C and D).
CDKN2B-AS1 Targeting miR-424-5p to Regulate HCC Cell Viability
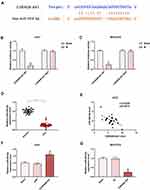
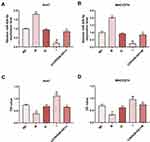
The target gene of CDKN2B-AS1 was predicted to be miR-424-5p by Starbase and further verified by dual-luciferase assay (Figure 5A). For both Huh7 and MHCC97H cells transfected with CDKN2B-AS1-WT, luciferase activities of the two cells in the mimic group were significantly lower than those in the blank group (Figure 5B and C), which confirmed that CDKN2B-AS1 targeted miR-424-5p. In addition, in our 30 HCC tissue samples, the expression of miR-424-5p was lower than that in normal tissues (Figure 5D), we found that expression levels of CDKN2B-AS1 were negatively correlated with those of miR-424-5p (Figure 5E). The expression of miR-424-5p in Huh7 transfected with siCDKN2B-AS1 and MHCC97H transfected with CDKN2B-AS1 was measured (Figure 5F and G), the results demonstrated that miR-424-5p was high-expressed when the expression of CDKN2B-AS1 was down-regulated, but low-expressed when CDKN2B-AS1 was overexpressed. Next, Huh7 and MHCC97H cells were, respectively, transfected with siCDKN2B-AS1+miR-424-5p inhibitor and CDKN2B-AS1+miR-424-5p mimic, and their transfection efficiencies were detected (Figure 6A and B); moreover, the effects of CDKN2B-AS1 and miR-424-5p on the viabilities of the two cells were explored. The data showed that overexpression of miR-424-5p reduced the viabilities of the two cells, but miR-424-5p knockdown increased the viabilities of the two cells (Figure 6C and D). In addition, the cell viability of siCDKN2B-AS1+miR-424-5p inhibitor group was lower than that of the miR-424-5p inhibitor group in Huh7 cells, whereas the cell viability of CDKN2B-AS1+miR-424-5p mimic was higher than that of miR-424-5p mimic group in MHCC97H cells (Figure 6C and D).
Effects of miR-424-5p in Combination with CDKN2B-AS1 on Migration, Invasion and EMT of HCC Cells
The miR-424-5p mimic and miR-424-5p inhibitor were transfected into Huh7 and MHCC97H cells, respectively, and the wound-healing and Transwell assay were performed to detect cell migration and invasion (Figure 7). The results showed that overexpression of miR-424-5p significantly reduced cell migration and invasion of Huh7 and MHCC97H cells, which were increased by miR-424-5p knockdown. Moreover, siCDKN2B-AS1 and miR-424-5p inhibitor were transfected into Huh7 cells and CDKN2B-AS1 vector and miR-424-5p mimic were transfected into MHCC97H cells. The results showed that in the presence of miR-424-5p inhibitor, siCDKN2B-AS1 significantly reduced cell migration and invasion of Huh7 cells, while in the presence of up-regulation of miR-424-5p, the overexpression of CDKN2B-AS1 significantly increased cell migration and invasion of MHCC97H cells compared the MHCC97H cells only transfected with miR-424-5p mimic. Moreover, the expressions of proteins related to EMT were examined by Western blot (Figure 8), we observed that the expression of E-Cadherin was high and the expressions of N-Cadherin and Snail were low in the cell groups of miR-424-5p mimic, while completely opposite results were detected in the cells transfected with miR-424-5p inhibitor. In addition, the E-Cadherin level was higher and N-Cadherin and Snail levels were lower in the cells with silencing CDKN2B-AS1 and miR-424-5p inhibitor than in the cells with miR-424-5p inhibitor. However, E-cadherin level was lower and N-Cadherin and Snail levels were higher in the cells with up-regulated CDKN2B-AS1 and miR-424-5p mimic compared with the cells with miR-424-5p mimic.
Discussion
Highly metastatic nature of HCC has attracted much research attention for the investigation of its metastatic mechanisms. Studying tumorigenesis mechanism of HCC helps discover the potential tumor suppressors, improve the treatment and prognosis of HCC. Cancer occurrence refers to the process during which normal cells develop into tumors, and malignant cells derive from normal human cells with the changes of transcriptome inside the cells.20
The potential values of lncRNAs in promoting or suppressing tumor growth have been recognized. In the present study, lncRNA CDKN2B-AS1 was selected as subject. CDKN2B anti-sense RNA (CDKN2B-AS1) is located within the CDKN2B-CDKN2A gene cluster at chromosome 9p21.21 CDKN2B participates in the development of many diseases such as coronary heart disease (CHD),22 myocardial infarction,23 stroke,24 brain disease,25 and inflammatory bowel disease in Koreans.26 The association between CDKN2B-AS1 and cancers is less explored. In cervical cancer cells, the interference of CDKN2B-AS1 suppresses the metastasis and activates cell apoptosis and senescence-related proteins;19 CDKN2B-AS1 promotes tumor growth of ovarian cancer cells via sponging miR-411-3p in vitro and HIF/1a/VEGF/P38 signaling pathway in vivo;27 CDKN2B-AS1 expression is observed increased in HCC tissues and cell lines. MHCC97H cells with a high-expressed CDKN2B-AS1 showed increased cell viability, and more aggressive migration and invasion, while down-regulation of CDKN2B-AS1 in Huh7 cells resulted in a lower viability, and inhibited cell migration and invasion. Furthermore, the expression of CDKN2B-AS1 also affects EMT of HCC cells. EMT is a biological process through which epithelial cells transform into mesenchymal phenotypic cells, and it plays an irreplaceable role in oncoma metastasis. Once EMT process was initiated, epithelial cells lose attachments and tight junctions from other cells and gradually obtain mesenchymal abilities that allow them to detach from membrane barriers of tissues.28 Moreover, mesenchymal phenotypic cells enhance migration and invasion, anti-apoptosis, and extracellular matrix degradation of mesenchymal phenotypic cells.29 EMT is also characterized by decreased E-Cadherin and increased N-Cadherin expression level.30 Snail family encodes transcriptional repressors31 and promotes cancer initiation and development. Snail could increase cell migration and invasion through repressing epithelial markers and promoting expressions of mesenchymal markers.32 The current study showed that silencing CDKN2B-AS1 increased E-Cadherin expression and reduced N-Cadherin and Snail expressions, whereas the overexpression of CDKN2B-AS1 resulted in opposite results, suggesting that CDKN2B-AS1 was closely related with the occurrence of EMT.
LncRNAs could regulate some other RNA transcripts by targeting specific miRNAs.33 Study found that lncRNA SNHG20 takes part in cell proliferation, invasion, and migration by regulating miR-495 in breast cancer;34 through acting as miR-497 sponger, lnc-SNHG1 is involved in the promotion of non-small cell lung cancer initiation and development;35 lncNR2C2-uORF targets UCA1-miR-627-5p-NR2C2 feedback loop to further modulate the malignant behaviors of glioma cells.36 In the current study, starBase predicted that the target gene of CDKN2B-AS1 was miR-424-5p. MiR-424-5p locates in human chromosome Xq26.3 and is in a cluster with miR-15/miR-16.37 The results of current study demonstrated that overexpression of miR-424-5p reduced cell viability, inhibited cell migration and invasion capacities, and also affected EMT, while the inhibition of miR-424-5p expression promoted cell viability, migration, invasion, and EMT process. Moreover, silencing miR-424-5p reversed the viability, migration and invasion capabilities and EMT process previously inhibited by CDKN2B-AS1 knockdown. However, the overexpression of miR-424-5p suppressed the high cell viability, migration, and invasion, and promoted EMT previously caused by up-regulation of CDKN2B-AS1. There are a few studies investigated the role of miR-424-5p in cancers and other diseases. In muscle wasting, miR-424-5p suppresses synthesis of ribosomal RNAs and proteins;38 miR-424-5p inhibits the metastasis and invasion of intrahepatic cholangiocarcinoma via sponging ARK5;39 in HCC, miR-424-5p reverses EMT and inhibits HCC progression via directly sponging ICAT.40
Conclusions
In conclusion, the present study investigated the relationship between lncRNA CDKN2B-AS1 and cell viability, migration, invasion, and EMT of HCC cells, and we found that CDKN2B-AS1 could promote HCC progression by increasing cell viability, migration, and invasion and promoting EMT process via targeting miR-424-5p. The current findings provide a possible target for HCC treatment.
Abbreviations
HCC, hepatocellular carcinoma; CDKN2B-AS1, CDKN2B antisense RNA 1; CHD, coronary heart disease; EMT, epithelial–mesenchymal transition.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. No animals are involved in this research.
Disclosure
The athors declare no conflicts of interest.
References
1. Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi:10.3322/canjclin.57.1.43
2. Desai JR, Ochoa S, Prins PA, He AR. Systemic therapy for advanced hepatocellular carcinoma: an update. J Gastrointest Oncol. 2017;8(2):243–255. doi:10.21037/jgo.2017.02.01
3. Tang ZY, Ye SL, Liu YK, et al. A decade’s studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130(4):187–196. doi:10.1007/s00432-003-0511-1
4. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
5. Soudyab M, Iranpour M, Ghafouri-Fard S. The role of long non-coding RNAs in breast cancer. Arch Iran Med. 2016;19(7):508–517.
6. Wu ZY, Wang SG, Li Q, Zhao QS, Shao MM. Identification of 10 differently expressed lncRNAs as prognostic biomarkers for prostate adenocarcinoma. Math Biosci Eng. 2019;17(3):2037–2047. doi:10.3934/mbe.2020108
7. Zhang C, Lian H, Xie L, Yin N, Cui Y. LncRNA ELFN1-AS1 promotes esophageal cancer progression by up-regulating GFPT1 via sponging miR-183-3p. Biol Chem. 2019. doi:10.1515/hsz-2019-0430
8. Lei K, Liang X, Gao Y, et al. Lnc-ATB contributes to gastric cancer growth through a MiR-141-3p/TGFbeta2 feedback loop. Biochem Biophys Res Commun. 2017;484(3):514–521. doi:10.1016/j.bbrc.2017.01.094
9. Liu F, Hu L, Pei Y, et al. Long non-coding RNA AFAP1-AS1 accelerates the progression of melanoma by targeting miR-653-5p/RAI14 axis. BMC Cancer. 2020;20(1):258. doi:10.1186/s12885-020-6665-2
10. Huo X, Han S, Wu G, et al. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16(1):165. doi:10.1186/s12943-017-0734-4
11. Jiang R, Tang J, Chen Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129. doi:10.1038/ncomms15129
12. You G, Zhou C, Xuan W. LncRNA LINC00668 promotes cell proliferation, migration, invasion ability and EMT process in hepatocellular carcinoma by targeting miR-532-5p/YY1 axis. Biosci Rep. 2020;40(5).
13. Shan Z, Wei L, Yu S, et al. Ketamine induces reactive oxygen species and enhances autophagy in SV-HUC-1 human uroepithelial cells. J Cell Physiol. 2019;234(3):2778–2787. doi:10.1002/jcp.27094
14. Zhang Y, Tang B, Song J, et al. Lnc-PDZD7 contributes to stemness properties and chemosensitivity in hepatocellular carcinoma through EZH2-mediated ATOH8 transcriptional repression. J Exp Clin Cancer Res. 2019;38(1):92. doi:10.1186/s13046-019-1106-2
15. Lasek-Bal A, Kula D, Urbanek T, et al. The association of SNPs located in the CDKN2B-AS1 and LPA genes with carotid artery stenosis and atherogenic stroke. Front Neurol. 2019;10:1170. doi:10.3389/fneur.2019.01170
16. Liu F, Xiao Y, Ma L, Wang J. Regulating of cell cycle progression by the lncRNA CDKN2B-AS1/miR-324-5p/ROCK1 axis in laryngeal squamous cell cancer. Int J Biol Markers. 2020;35(1):47–56. doi:10.1177/1724600819898489
17. Gui D, Cao H. Long non-coding RNA CDKN2B-AS1 promotes osteosarcoma by increasing the expression of MAP3K3 via sponging miR-4458. In Vitro Cell Dev Biol Anim. 2020;56(1):24–33. doi:10.1007/s11626-019-00415-7
18. Cui X, Yu T, Shang J, Xiao D, Wang X. Long non-coding RNA CDKN2B-AS1 facilitates laryngeal squamous cell cancer through regulating miR-497/CDK6 pathway. Onco Targets Ther. 2019;12:8853–8862. doi:10.2147/OTT.S221620
19. Zhu L, Zhang Q, Li S, Jiang S, Cui J, Dang G. Interference of the long noncoding RNA CDKN2B-AS1 upregulates miR-181a-5p/TGFbetaI axis to restrain the metastasis and promote apoptosis and senescence of cervical cancer cells. Cancer Med. 2019;8(4):1721–1730. doi:10.1002/cam4.2040
20. Balani S, Nguyen LV, Eaves CJ. Modeling the process of human tumorigenesis. Nat Commun. 2017;8(1):15422. doi:10.1038/ncomms15422
21. Jarinova O, Stewart AF, Roberts R, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29(10):1671–1677. doi:10.1161/ATVBAHA.109.189522
22. Dehghan A, Bis JC, White CC, et al. Genome-wide association study for incident myocardial infarction and coronary heart disease in prospective cohort studies: the CHARGE consortium. PLoS One. 2016;11(3):e0144997. doi:10.1371/journal.pone.0144997
23. Matsuoka R, Abe S, Tokoro F, et al. Association of six genetic variants with myocardial infarction. Int J Mol Med. 2015;35(5):1451–1459. doi:10.3892/ijmm.2015.2115
24. Bai Y, Nie S, Jiang G, et al. Regulation of CARD8 expression by ANRIL and association of CARD8 single nucleotide polymorphism rs2043211 (p.C10X) with ischemic stroke. Stroke. 2014;45(2):383–388. doi:10.1161/STROKEAHA.113.003393
25. Zhao J, Wu X, Nie S, et al. Association of CDKN2B-AS1 rs1333049 with brain diseases: a case-control study and a meta-analysis. Clin Psychopharmacol Neurosci. 2017;15(1):53–58. doi:10.9758/cpn.2017.15.1.53
26. Lee HS, Lee SB, Kim BM, et al. Association of CDKN2A/CDKN2B with inflammatory bowel disease in Koreans. J Gastroenterol Hepatol. 2018;33(4):887–893. doi:10.1111/jgh.14031
27. Wang Y, Huang Y, Liu H, Su D, Luo F, Zhou F. Long noncoding RNA CDKN2B-AS1 interacts with miR-411-3p to regulate ovarian cancer in vitro and in vivo through HIF-1a/VEGF/P38 pathway. Biochem Biophys Res Commun. 2019;514(1):44–50. doi:10.1016/j.bbrc.2019.03.141
28. Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112(12):1776–1784. doi:10.1172/JCI200320530
29. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi:10.1038/nrm3758
30. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
31. Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3(3):155–166. doi:10.1038/nrm757
32. Peinado H, Olmeda D, Cano A. Snail, zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi:10.1038/nrc2131
33. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
34. Guan YX, Zhang MZ, Chen XZ, Zhang Q, Liu SZ, Zhang YL. Lnc RNA SNHG20 participated in proliferation, invasion, and migration of breast cancer cells via miR-495. J Cell Biochem. 2018;119(10):7971–7981. doi:10.1002/jcb.26588
35. Li Z, Lu Q, Zhu D, Han Y, Zhou X, Ren T. Lnc-SNHG1 may promote the progression of non-small cell lung cancer by acting as a sponge of miR-497. Biochem Biophys Res Commun. 2018;506(3):632–640. doi:10.1016/j.bbrc.2018.10.086
36. Fan Z, Zheng J, Xue Y, et al. NR2C2-uORF targeting UCA1-miR-627-5p-NR2C2 feedback loop to regulate the malignant behaviors of glioma cells. Cell Death Dis. 2018;9(12):1165. doi:10.1038/s41419-018-1149-x
37. Chamorro-Jorganes A, Araldi E, Penalva LO, Sandhu D, Fernandez-Hernando C, Suarez Y. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol. 2011;31(11):2595–2606. doi:10.1161/ATVBAHA.111.236521
38. Connolly M, Paul R, Farre-Garros R, et al. miR-424-5p reduces ribosomal RNA and protein synthesis in muscle wasting. J Cachexia Sarcopenia Muscle. 2018;9(2):400–416. doi:10.1002/jcsm.12266
39. Wu J, Yang B, Zhang Y, et al. miR-424-5p represses the metastasis and invasion of intrahepatic cholangiocarcinoma by targeting ARK5. Int J Biol Sci. 2019;15(8):1591–1599. doi:10.7150/ijbs.34113
40. Zhang Y, Li T, Guo P, et al. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep. 2014;4:6248. doi:10.1038/srep06248
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.