Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA-ATB Promotes the Tumorigenesis of Ovarian Cancer via Targeting miR-204-3p
Authors Yuan D, Qian H, Guo T, Ye J, Jin C, Liu X, Jiang L, Wang X, Lin M, Yu H
Received 10 September 2019
Accepted for publication 22 December 2019
Published 20 January 2020 Volume 2020:13 Pages 573—583
DOI https://doi.org/10.2147/OTT.S230552
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Donglan Yuan, 1,* Hua Qian, 1,* Ting Guo, 2 Jun Ye, 2 Chunyan Jin, 2 Xia Liu, 1 Li Jiang, 1 Xiaoxiang Wang, 1 Mei Lin, 3 Hong Yu 4
1Department of Obstetrics and Gynecology, Taizhou People’s Hospital, Taizhou, Jiangsu, People’s Republic of China; 2Center for Molecular Medicine, Taizhou People’s Hospital, Taizhou, Jiangsu, People’s Republic of China; 3Scientific Research Office, Taizhou People’s Hospital, Taizhou, Jiangsu, People’s Republic of China; 4Department of Pathology, Taizhou People’s Hospital, Taizhou, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hong Yu
Department of Pathology, Taizhou People’s Hospital, 399 Hailing Road, Taizhou, Jiangsu 225300, People’s Republic of China
Email [email protected]
Background: Ovarian cancer ranks fifth among the most prevalent cancer type in females all over the world. It is the second most frequent malignant tumor which accounts for 3% of cancer in females. Therefore, to explore the mechanism of carcinogenesis in ovarian cancer is important to develop new treatment methods. It has been previously found that lncRNA-ATB could promote the tumorigenesis of malignant tumors. However, the role of lncRNA-ATB during the progression of ovarian cancer remains unclear.
Methods: Gene expressions in tissues or cells were detected by using qRT-PCR. Western blot was performed to investigate the protein expressions in ovarian cancer cells. Cell apoptosis was tested by flow cytometry. Moreover, the correction between lncRNA-ATB and miR-204-3p was examined by Dual-luciferase reporter assay and RNA pulldown. Cell proliferation and invasion were detected by CCK-8, Ki-67 staining and transwell assay, respectively. Finally, xenograft mice model was established to confirm the result of in vitro experiments.
Results: LncRNA-ATB silencing significantly inhibited the proliferation and induced apoptosis of ovarian cancer cells. In addition, luciferase activity suggested that lncRNA-ATB negatively regulated miR-204-3p in ovarian cancer. Besides, Nidogen 1 (NID1) was the direct target of miR-204-3p. Overexpression of NID1 could notably reverse the inhibitory effect of lncRNA-ATB knockdown on the progression of ovarian cancer. Finally, lncRNA-ATB silencing notably attenuated the severity of ovarian cancer in vivo.
Conclusion: Downregulation of lncRNA-ATB significantly inhibited the tumorigenesis of ovarian cancer in vitro and in vivo, which may serve as a potential novel target for the treatment of ovarian cancer.
Keywords: ovarian cancer, lncRNA-ATB, miR-204-3p, NID1
Introduction
Ovarian cancer is presently the fifth leading cause of death among the most prevalent cancer type in females all over the world.1 Several living habits and environmental factors have been confirmed to contribute to the progression of ovarian cancer.2 Although some efforts have been made in understanding the pathogenesis of ovarian cancer, effective strategies are still limited in decreasing the incidence and recurrence rate related to ovarian cancer.3 Therefore, it is necessary to find novel mechanisms of tumorigenesis in ovarian cancer.
A large number of reports have considered noncoding RNAs (ncRNAs) as possible regulators of multiple diseases.4–6 MicroRNAs are small ncRNAs with a length of 20-25nt that are involved in the pathogenesis of ovarian cancer.7 These ncRNAs are greater than 200 nucleotides in length with limited or no protein-coding capacity, and they are known as long non-coding RNAs (lncRNAs).8 LncRNAs have been regarded to play a key role in tumorigenesis of ovarian cancer. For instance, Yan et al found that lncRNA FLVCR1-AS1 could promote the cell proliferation and invasion of ovarian cancer.9 Additionally, You et al indicated that lincRNA DLX6-AS1 could act as an oncogene by targeting miR-613 in ovarian cancer.10 Previous studies have indicated that lncRNA-ATB could significantly promote the progression of multiple malignancies.11–13 However, the role of lncRNA-ATB in the progression of ovarian cancer remains to be explored. Thus, this research aimed to explore the biological function of lncRNA-ATB during the progression of ovarian cancer in vitro and in vivo.
Materials and Methods
Cell Culture
SKOV3 A2780 and 293T cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in RPMI-1640 medium, supplemented with 10% FBS and 2 mM Glutamine (Sigma-Aldrich, St. Louis, MO, USA) at 37°C.
Cell Transfection
Lentiviral expressing short-hairpin RNA (shRNA1 or shRNA2) directed target lncRNA-ATB and one nontargeting sequence (negative control) were obtained from Hanbio Biotechnology Co., Ltd (Shanghai, China). Next, lncRNA-ATB shRNA1 or shRNA2 was packaged into lentiviruses. Then, the lentiviral vector DNAs were then transfected into 293T cells including lenti-lncRNA-ATB shRNAs and negative control (NC). After transfection, the cells were incubated at 32°C, and then the supernatant was collected. After that, supernatants of two lncRNA-ATB shRNAs and negative control were filtered into particles. Finally, all ovarian cancer cells were infected with lentiviral particles according to the manufactures’ protocol. After 48 h of incubation, stable ovarian cancer cells were then selected by puromycin (2.5 μg/mL, Sigma Aldrich, St. Louis, MO, USA). qRT-PCR assay was used to verify the efficiency of transfection.
For miR-204-3p transfection, SKOV3 cells were transfected with miR-204-3p inhibitor or NC by Lipofectamine 2000 according to the previous reference.14 MiR-204-3p inhibitor and negative control RNAs were purchased from GenePharma (Shanghai, China). The efficiency of transfection was detected by q-PCR.
NID1 Overexpression
SKOV3 or A2780 cells were plated into 60-mm plates at 4×105 cells/well overnight. Then, supernatants with lentiviruses carrying the NID1 gene were added directly to cancer cells (at 50–60% of confluence) for 24 hrs. Next, SKOV3 cells were re-plated on the selection medium-containing puromycin (2.5 μg/mL) for another 3 days.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNAs were extracted from tissues or cell lines with TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). We carried out reverse transcription and real-time PCR assays by means of PrimeScript RT reagent Kit (Takara, Tokyo, Japan) and SYBR premix Ex Taq II kit (Takara, Tokyo, Japan) severally. β-actin or U6 was used as the internal control. The primers for lncRNA-ATB were: forward: 5′-GAGGCTGGTTGACATGCCTT-3ʹ and reverse: 5′-GAGCATCTCTGGGTGCTGGT-3′; the primers for NID1 were: forward: 5′-CCTTCATAACTGCGACATACCC-3′ and reverse: 5′-AAGCCGTCTCCCTGATAACC-3′; the primers for miR-204-3p were: forward: 5′-GGGAAGGCAAAGGGACGT-3′ and reverse: CTCAACTGGTGTCGTGGATGC; the primers for U6 were: forward: CTCGCTTCGGCAGCACAT and reverse: AACGCTTCACGAATTTGCGT and the primers for β-actin were: forward: 5′-GTCCACCGCAAATGCTTCTA-3′ and reverse: 5′-TGCTGTCACCTTCACCGTTC-3′. 2−ΔΔCT method was utilized to measure the relative expression.
CCK-8 Assay
Cell counting kit-8 assay (CCK8, Beyotime, Shanghai, China) was used for testing the cell viability. SKOV3 or A2780 cells were plated into 96-well plates with a density of 5×103 cells per well and treated as following: nothing (Blank), lentiviral-control (NC), lncRNA-ATB shRNA2 (lncRNA-ATB shRNA2) or lncRNA-ATB shRNA2 plus miR-204-3p inhibitor (lncRNA-ATB shRNA2+miR-204-3p inhibitor) for 0, 24, 48 and 72 hrs, respectively. Then, ovarian cancer cells were treated with 10-μL CCK-8 reagent for another 2 h at 37°C. Next, the absorbance of cells was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
RNA Pulldown
For the RNA pulldown assay, the Biotin RNA Labeling Mix (Roche, Basel, Switzerland) was used to transcribe and label probe-control or probe-ATB from ATB shRNA lenti vector in vitro. An RNA structure buffer (Thermo, MA, USA) was used to induce secondary structure formation from the biotin-labeled RNAs. Streptavidin beads (Thermo) were washed three times with 500 μL of RNA immunoprecipitation wash buffer (Thermo) and then added to the biotinylated RNAs at 4°C overnight. The overnight mixture was separated by a magnetic field so that streptavidin bead-RNA complexes could be obtained. Then, lysates of SKOV3 cells were added to the complexes and incubated on a rotator at room temperature for 1 hr. The incubated mixture was again separated with a magnetic field so that streptavidin bead-RNA-protein complexes could be obtained.
Western Blotting
Total proteins were lysed using RIPA lysis buffer. Then, the concentration of protein was detected using a BCA protein kit (Thermo Fisher Scientific, Waltham, MA, USA). Proteins (40 μg per lane) were separated on 10% SDS-PAGE gel and then transferred onto polyvinylidene fluoride (PVDF, Thermo Fisher Scientific) membranes. After that, the membranes were blocked with 5% skim milk in TBST for 1 h at room temperature, and then incubated with the primary antibodies against NID1 (Abcam Cambridge, MA, USA, 1:1000), Bax (Abcam, 1:1000), Bcl-2 (Abcam, 1:1000), cleaved caspase 3 (Abcam, 1:1000), E-cadherin (Abcam, 1:1000), vimentin (Abcam, 1:1000) and β-actin (Abcam, 1:1000) overnight at 4°C. Then, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Finally, the membranes were detected by Enhanced Chemiluminescence (ECL) kit (Thermo Fisher Scientific, Waltham, MA, USA). β-actin was used as an internal control.
Immunofluorescence
Ovarian cancer cells or tumor tissues of mice were prefixed in 4% paraform for 10 mins, and fixed in pre-cold methanol for another 10 mins. Next, cells were incubated with primary antibodies overnight at 4°C: anti-Ki67 (Abcam; 1:1000) and DAPI (Abcam; 1:1000). Goat anti-rabbit IgG antibody (Abcam; 1:5000) was used as the secondary antibody. The samples were visualized by fluorescence microscope (Olympus CX23, Tokyo, Japan) immediately.
Cell Apoptosis Analysis
SKOV3 or A2780 cells were seeded in a 6-well plate. The residue was resuspended with 100-μL binding buffer after centrifuged at 1000 rpm/min for 5 min. Then, 5-μL Annexin V-FITC and 5-μL propidium (PI) were added in the system for 15 min. The cell apoptotic rate was measured by flow cytometer (BD, Franklin Lake, NJ, USA) and the results were analyzed using the software WinMDI 2.9 (Invitrogen, Carlsbad, CA, USA).
Dual-Luciferase Reporter Assay
The partial sequences of lncRNA-ATB and 3′-UTR of NID1 containing the putative binding sites of miR-204-3p were synthetized and obtained from Sangon Biotech (Shanghai, China), then were cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vectors (Promega, Madison, WI, USA) to construct wild-type reporter vectors ATB (WT) and NID1 (WT), respectively. The mutant lncRNA-ATB sequences and 3′-UTR of NID1 sequences containing the putative binding sites of miR-204-3p were performed by Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA, USA) and then cloned into pmirGLO vectors, respectively, to construct mutant-type reporter vectors ATB (MUT) and NID1 (MUT). The ATB (WT) or ATB (MUT) were transfected into SKOV3 cells together with Blank, vector-control or miR-204-3p mimics using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Similarly, the NID1 (WT) or NID1 (MUT) was transfected into 293T cells together with Blank, vector-control or miR-204-3p mimics. The relative luciferase activity was analyzed by the Dual-Glo Luciferase Assay System (Promega, Madison, WI, USA).
Transwell Assay
For cell invasion analysis, transwell assay was performed in this study. The upper chamber is pre-treated with 100 μL of Matrigel. SKOV3 or A2780 cells were seeded into the upper chamber in media with 1% FBS, and the density was adjusted to about 1.0×106 cells per chamber. RPMI1640 medium with 10% FBS was added to the lower chamber. After 24 hrs of incubation at 37°C, the transwell chamber was rinsed twice with PBS (5 min per time), fixed by 5% glutaraldehyde at 4°C and stained with 0.1% crystal violet for 30 mins. The transwell chamber was washed twice with PBS and then observed under a microscope. The number of cells invading the Matrigel was regarded to be a reflection of the invasion ability.
In vivo Experiments
9 BALB/nude mice (aged 6 weeks) were purchased from Vital River (Beijing, China) and housed within a dedicated SPF facility. The nude mice were injected subcutaneously with SKOV3 cells (5×106 cells, in 100 μL of PBS) into the right flanks of mice according to the previous reference.15 LncRNA-ATB shRNA2 or vector control was directly injected into the mice. Tumor size was measured weekly for 4 weeks according to the equation: (length×width^2)/2. At the end of the experiments, the mice were sacrificed for the collection of tumors. Then, each tumor was weighed. All in vivo experiments were performed in accordance with National Institutes of Health guide for the care and use of laboratory animals, following a protocol approved by the Ethics Committees of Taizhou People’s Hospital.
Statistical Analysis
Each group were performed at least three independent experiments and all data were expressed as the mean ± standard deviation (SD). The comparison between the two groups was analyzed by Student’s t-test. The comparisons among multiple groups were made with one-way analysis of variance (ANOVA) followed by Tukey’s test (Graphpad Prism7). P<0.05 was considered to indicate a statistically significant difference.
Results
ATB Silencing Inhibited the Proliferation of Ovarian Cells
To investigate the efficacy of transfections, the expression of ATB in ovarian cancer cells were measured with q-PCR. As revealed in Figure 1A, lncRNA-ATB shRNA1 or shRNA2 significantly decreased the expression of lncRNA-ATB in ovarian cancer cells. Since lncRNA-ATB shRNA2 exhibited better silencing effect, it was used for the following experiments. Next, CCK-8 and Ki-67 staining were used to investigate the proliferation of ovarian cancer cells. As shown in Figure 1B and C, the cell viability of SKOV3 or A2780 cells was significantly decreased in the presence of lncRNA-ATB silencing, compared with control. Then, Ki-67 positive cell rate of SKOV3 was significantly decreased by lncRNA-ATB shRNA2 (Figure 1D and E). All these data demonstrated that lncRNA-ATB silencing significantly inhibited the proliferation of ovarian cancer cells.
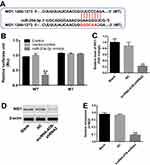
miR-204-3p Was the Downstream Target Gene of lncRNA-ATB
Next, to investigate the potential mechanism by which lncRNA-ATB silencing inhibited the growth of ovarian cancer in vitro, TargetScan and miRDB database blast were performed to search for the downstream target gene of lncRNA-ATB. As indicated in Figure 2A and B, lncRNA-ATB had a putative miR-204-3p targeting site. In addition, Luciferase reporter assay was performed to determine whether miR-204-3p could directly interact with lncRNA-ATB in SKOV3 cells. The result indicated that the co-transfection of the wild-type lncRNA-ATB vector (WT- lncRNA-ATB) with miR-204-3p mimics, significantly reduced luciferase activities compared with mutant lncRNA-ATB vector (MT- lncRNA-ATB) (Figure 2C). Similarly, the results of RNA pulldown demonstrated that ATB bound to miR-204-3p (Figure 2D). All these data showed that miR-204-3p was the downstream target gene of lncRNA-ATB.
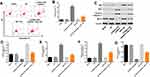
miR-204-3p Partially Reversed the Anti-Tumor Effect of lncRNA-ATB Silencing
In order to verify the transfection efficiency, q-PCR was used. As revealed in Figure 3A, miR-204-3p was downregulated in SKOV3 cells in the presence of miR-204-3p inhibitor. These data suggested that miR-204-3p was stably transfected into SKOV3 cells. Moreover, the downregulation of miR-204-3p significantly rescued the anti-proliferative and apoptotic effect of lncRNA-ATB silencing on ovarian cancer in vitro (Figure 3B–D). Taken together, MiR-204-3p partially reversed the anti-tumor effect of lncRNA-ATB silencing.
NID1 Was the Direct Target of miR-204-3p
Then, we applied TargetScan and miRDB database blast to explore the direct target of miR-204-3p. As indicated in Figure 4A, NID1 might be a potential target of miR-204-3p. In addition, the luciferase assay data indicated that reduced luciferase activity was observed in SKOV3 cells following transfection with WT-NID1 and miR-204-3p mimics (Figure 4B). These data indicated that NID1 was the direct target of miR-204-3p. Then, to explore the relation between lncRNA-ATB and NID1, q-PCR and Western blot were detected. As revealed in Figure 4C–E, the expression of NID1 in SKOV3 cells was significantly inhibited in the presence of lncRNA-ATB silencing. These results revealed that lncRNA-ATB indirectly targeted NID1.
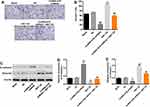
LncRNA-ATB Silencing Notably Induced the Apoptosis of SKOV3 Cells
In order to investigate the role of lncRNA-ATB during the tumorigenesis of ovarian cancer, flow cytometry was performed. As showed in Figure 5A and B, the apoptosis rate of SKOV3 cells was significantly increased by lncRNA-ATB knockdown. However, NID1 overexpression partially reversed the apoptotic effect of lncRNA-ATB silencing on ovarian cancer cells. Then, western blot was used to detect the expression of NID1 and apoptosis-related proteins. As illustrated in Figure 5C and D, the expressions of NID1in SKOV3 cells were notably decreased by lncRNA-ATB knockdown, which was partially rescued in the presence of NID1 overexpression. In contrast, the downregulation of lncRNA-ATB obviously increased the expression of pro-apoptosis proteins (Bax2 and Cleaved caspase3). However, the apoptotic effect of lncRNA-ATB knockdown on ovarian cancer cells was reversed by the upregulation of NID1 (Figure 5C, E and F). Besides, the anti-apoptotic protein (Bcl-2) in ovarian cancer cells was notably inactivated by knockdown of lncRNA-ATB, which was partially rescued in the presence of NID1 OE (Figure 5C and G). Taken together, all these results showed that lncRNA-ATB silencing could induce the apoptosis of ovarian cancer cells via activating pro-apoptosis proteins.
LncRNA-ATB Knockdown Notably Inhibited the Invasion of Ovarian Cancer Cells
Afterwards, transwell assay was performed to detect the effect of lncRNA-ATB on the invasion of ovarian cancer cells. As indicated in Figure 6A and B, the invasion cell number in SKOV3 cells was notably decreased by the downregulation of lncRNA-ATB, which was partially reversed by overexpression of NID1. Furthermore, overexpression of NID1 exhibited a significantly invasive effect on ovarian cancer cells. Then, the expressions of EMT-related proteins were detected by Western blot. The results showed that the downregulation of lncRNA-ATB significantly upregulated the expression of E-cadherin. However, the promoting effect of lncRNA-ATB silencing was partially rescued by the upregulation of NID1. In addition, NID1 OE alone notably inhibited the expression of E-cadherin (Figure 6C and D). In contrast, the expression of vimentin in ovarian cancer cells was obviously decreased in lncRNA-ATB shRNA2 but upregulated in NID1 OE group. Moreover, NID1 overexpression partially reversed the effect of lncRNA-ATB silencing on this protein (Figure 6C and E). Altogether, lncRNA-ATB knockdown could inhibit the invasion of ovarian cancer cells.
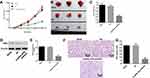
LncRNA-ATB Silencing Ameliorated the Severity of Ovarian Cancer in vivo
For the aim to investigate the effect of lncRNA-ATB on ovarian cancer in vivo, xenograft mice model was established. As demonstrated in Figure 7A and B, the tumor sizes in mice were significantly decreased by lncRNA-ATB shRNA2, compared with control. Similarly, lncRNA-ATB knockdown notably decreased the tumor weights of mice (Figure 7C). Moreover, the expression of NID1 in tumor tissues of mice was significantly decreased in the presence of lncRNA-ATB silencing (Figure 7D and E). Besides, the Ki67 positive rate of tumor tissues was obviously decreased by knockdown of lncRNA-ATB (Figure 7F and G). All these data revealed that lncRNA-ATB knockdown obviously ameliorated the symptom of ovarian cancer in vivo.
Downregulation of lncRNA-ATB Notably Inhibited the Progression of Ovarian Cancer in vitro
Finally, to verify the function of lncRNA-ATB on the other ovarian cancer cell line (A2780 cells), Ki-67 staining was used. As demonstrated in Supplementary Figure 1A and B, the integrated optical density (IOD) of A2780 cells was obviously decreased in the presence of lncRNA-ATB shRNA2. Similarly, knockdown of lncRNA-ATB significantly induced the apoptosis of A2780 cells, while the apoptotic effect of lncRNA-ATB silencing was partially reversed by overexpression of NID1 (Supplementary Figure 1C and D). Moreover, the invasion of A2780 cells was greatly inhibited by lncRNA-ATB shRNA2, which was significantly rescued by NID1 OE (Supplementary Figure 1E and F). In summary, the downregulation of lncRNA-ATB notably inhibited the progression of ovarian cancer in vitro.
Discussion
Recent studies have indicated that lncRNAs played a key role in the progression of ovarian cancer.16,17 In the present research, we confirmed that the expression of lncRNA-ATB was significantly upregulated in ovarian cancer cells. Li et al found that the expression of lncRNA GAS5 was notably inhibited in ovarian cancer cells.18 This discrepancy might be due to the different biological function between lncRNAs. Some reports have indicated that lncRNA-ATB was involved in multiple malignant tumors. For instance, lncRNA-ATB could promote the proliferation, migration and invasion of cervical cancer or glioma.13,19 Our current study further confirmed these results, suggesting that lncRNA-ATB could act as an oncogene. In addition, our study firstly found that lncRNA-ATB played a critical role in the tumorigenesis of ovarian cancer, which supplemented the biological function of lncRNA-ATB.
Next, to further explore the biological role of lncRNA-ATB during the development of ovarian cancer, in vitro experiments were performed. We found that the downregulation of lncRNA-ATB could induce the apoptosis of ovarian cancer cells. Moreover, lncRNA-ATB silencing significantly upregulated the expression of Bax and cleaved caspase3 and inhibited the protein level of Bcl-2. Bax, cleaved caspase3 and Bcl-2 were key regulators of cell apoptosis.20,21 Bax and cleaved caspase3 have been regarded to be the pro-apoptosis protein during the cell apoptosis,22,23 while the upregulation of Bcl-2 could inhibit the apoptosis of cancer cells.23 These data were consistent with our research, indicating that lncRNA-ATB silencing induced the apoptosis of ovarian cancer cells via upregulating Bax and cleaved caspase3 and downregulating Bcl-2.
Besides, our findings indicated that lncRNA-ATB knockdown could notably inhibit the invasion of ovarian cancer cells. Previous studies have confirmed that epithelial‑mesenchymal transition (EMT) process played a key role during the invasion of cancer.24–26 Bhatti et al found that E-cadherin was involved in metastasis of malignant tumors.25 In addition, the expression of vimentin was upregulated during the invasion of cancer cells.27 These results were similar to our present data that lncRNA-ATB knockdown could notably inhibit the invasion of ovarian cancer through upregulating the expression of E-cadherin and downregulating the level of vimentin.
Then, we further investigated the mechanism by which lncRNA-ATB silencing inhibited the progression of ovarian cancer in vitro and in vivo. The result of Dual-luciferase reporter assay indicated that miR-204-3p was the downstream target gene of lncRNA-ATB. MiRNAs are highly conserved ncRNAs that exert versatile biological functions.28,29 Zhu et al indicated that LncRNA-ATB could promote viability, migration, and angiogenesis in human microvascular endothelial cells by sponging microRNA-195.30 In addition, lncRNA-ATB could promote the progression of colorectal cancer by regulating miR-200c/CDK2 axis.31 These findings were similar to our previous research, indicating that lncRNA-ATB promoted the tumorigenesis of ovarian cancer via sponging miR-204-3p.
It has been confirmed that miRNAs could exert their function mainly through binding their target genes.32,33 To explore the potential mechanism of miR-204-3p in the occurrence of ovarian cancer, TargetScan databases were applied to identify target genes of miR-204-3p. Our finding found Nidogen 1 (NID1) was a direct target of miR-204-3p, which has been regarded as an EMT regulator during the metastasis of cancer.34 Pedrola et al confirmed that NID1 could act as a novel target of ETV5 transcription factor which was involved in endometrial cancer invasion.35 MiR-192 has been regarded to inhibit the growth of cancer cell via targeting NID1.36 Our findings were consistent with these results indicating that lncRNA-ATB could promote the tumorigenesis of ovarian cancer via indirectly targeting NID1. Since PI3K/AKT/mTOR signaling pathway could be considered as a key regulator for ovarian cancer,37 we will further investigate the effect of lncRNA-ATB on PI3K/AKT/mTOR signaling in future.
In conclusion, lncRNA-ATB silencing could inhibit the tumorigenesis of ovarian cancer via regulating miR-204-3p/NID1 axis, which might serve as a potential novel target for treating ovarian cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kannan K, Coarfa C, Rajapakshe K, et al. CDKN2D-WDFY2 is a cancer-specific fusion gene recurrent in high-grade serous ovarian carcinoma. PLoS Genet. 2014;10(3):e1004216. doi:10.1371/journal.pgen.1004216
2. Mungenast F, Thalhammer T. Estrogen biosynthesis and action in ovarian cancer. Front Endocrinol (Lausanne). 2014;5:192. doi:10.3389/fendo.2014.00192
3. Kadry Taher M, Farhat N, Karyakina NA, et al. Critical review of the association between perineal use of talc powder and risk of ovarian cancer. Reprod Toxicol. 2019;90:88–101. doi:10.1016/j.reprotox.2019.08.015
4. Shen X, Zhu W. Long non-coding RNA LINC01627 is a prognostic risk factor for epithelial ovarian cancer. Oncol Lett. 2019;18(3):2861–2868. doi:10.3892/ol.2019.10661
5. Li N, Zhan X. Identification of clinical trait-related lncRNA and mRNA biomarkers with weighted gene co-expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA J. 2019;10(3):273–290. doi:10.1007/s13167-019-00175-0
6. Gong J, Xu X, Zhang X, Zhou Y. LncRNA MIR4435-2HG is a potential early diagnostic marker for ovarian carcinoma. Acta Biochim Biophys Sin (Shanghai). 2019;51:953–959. doi:10.1093/abbs/gmz085
7. Deb B, Uddin A, Chakraborty S. miRNAs and ovarian cancer: an overview. J Cell Physiol. 2018;233(5):3846–3854. doi:10.1002/jcp.v233.5
8. Frank S, Aguirre A, Hescheler J, Kurian L. A lncRNA perspective into (Re)Building the heart. Front Cell Dev Biol. 2016;4:128. doi:10.3389/fcell.2016.00128
9. Yan H, Li H, Silva MA, et al. LncRNA FLVCR1-AS1 mediates miR-513/YAP1 signaling to promote cell progression, migration, invasion and EMT process in ovarian cancer. J Exp Clin Cancer Res. 2019;38(1):356. doi:10.1186/s13046-019-1356-z
10. You Q, Shi HY, Gong CF, Tian XY, Li S. Long non-coding RNA DLX6-AS1 acts as an oncogene by targeting miR-613 in ovarian cancer. Eur Rev Med Pharmacol Sci. 2019;23(15):6429–6435. doi:10.26355/eurrev_201908_18524
11. Li RH, Chen M, Liu J, et al. Long noncoding RNA ATB promotes the epithelial-mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018;9(12):1171. doi:10.1038/s41419-018-1210-9
12. Song C, Xiong Y, Liao W, Meng L, Yang S. Long noncoding RNA ATB participates in the development of renal cell carcinoma by downregulating p53 via binding to DNMT1. J Cell Physiol. 2019;234(8):12910–12917. doi:10.1002/jcp.v234.8
13. Tang F, Wang H, Chen E, et al. LncRNA-ATB promotes TGF-beta-induced glioma cells invasion through NF-kappaB and P38/MAPK pathway. J Cell Physiol. 2019;234(12):23302–23314. doi:10.1002/jcp.28898
14. Shao X, Zhang S, Tang Y, Kong W. Micro RNA-30b (inhibitor) nanoparticles suppressed the lipopolysaccharide (LPS)-induced acute kidney injury. IET Nanobiotechnol. 2019;13(9):923–927. doi:10.1049/iet-nbt.2019.0110
15. Long X, Song K, Hu H, et al. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res. 2019;38(1):345. doi:10.1186/s13046-019-1329-2
16. Cai J, Li L, Song L, et al. Effects of long term antiprogestine mifepristone (RU486) exposure on sexually dimorphic lncRNA expression and gonadal masculinization in Nile tilapia (Oreochromis niloticus). Aquat Toxicol. 2019;215:105289. doi:10.1016/j.aquatox.2019.105289
17. Yan J, Jia Y, Chen H, Chen W, Zhou X. Long non-coding RNA PXN-AS1 suppresses pancreatic cancer progression by acting as a competing endogenous RNA of miR-3064 to upregulate PIP4K2B expression. J Exp Clin Cancer Res. 2019;38(1):390. doi:10.1186/s13046-019-1379-5
18. Li J, Yang C, Li Y, et al. LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep. 2017;38:BSR20171150.
19. Zhu Y, Wu Y, Yang L, et al. Long non-coding RNA activated by transforming growth factor-beta promotes proliferation and invasion of cervical cancer cells by regulating the miR-144/ITGA6 axis. Exp Physiol. 2019;104(6):837–844. doi:10.1113/EP087656
20. Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. doi:10.1155/2014/150845
21. Pan LL, Wang AY, Huang YQ, Luo Y, Ling M. Mangiferin induces apoptosis by regulating Bcl-2 and Bax expression in the CNE2 nasopharyngeal carcinoma cell line. Asian Pac J Cancer Prev. 2014;15(17):7065–7068. doi:10.7314/APJCP.2014.15.17.7065
22. Qi L, Jiang-Hua M, Ge-Liang H, Qing C, Ya-Ming L. MiR-34a inhibits spinal cord injury and blocks spinal cord neuron apoptosis by activating phatidylinositol 3-kinase (PI3K)/AKT pathway through targeting CD47. Curr Neurovasc Res. 2019. doi:10.2174/1567202616666190906102343
23. Raghav PK, Kumar R, Kumar V, Raghava GPS. Docking-based approach for identification of mutations that disrupt binding between Bcl-2 and Bax proteins: inducing apoptosis in cancer cells. Mol Genet Genomic Med. 2019;7:e910.
24. Bronsert P, Enderle-Ammour K, Bader M, et al. Cancer cell invasion and EMT marker expression: a three-dimensional study of the human cancer-host interface. J Pathol. 2014;234(3):410–422. doi:10.1002/path.2014.234.issue-3
25. Bhatti MZ, Pan L, Wang T, Shi P, Li L. REGgamma potentiates TGF-beta/Smad signal dependent epithelial-mesenchymal transition in thyroid cancer cells. Cell Signal. 2019;109412. doi:10.1016/j.cellsig.2019.109412
26. Zheng X, Carstens JL, Kim J, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi:10.1038/nature16064
27. Lin Z, Zhang L, Zhou J, Zheng J. Silencing Smad4 attenuates sensitivity of colorectal cancer cells to cetuximab by promoting epithelial‑mesenchymal transition. Mol Med Rep. 2019. doi:10.3892/mmr
28. Eguchi T, Kuboki T. Cellular reprogramming using defined factors and microRNAs. Stem Cells Int. 2016;2016:7530942. doi:10.1155/2016/7530942
29. Tam C, Wong JH, Tsui SKW, et al. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: updates in recent years. Appl Microbiol Biotechnol. 2019;103(12):4649–4677.
30. Zhu AD, Sun YY, Ma QJ, Xu F. lncRNA-ATB promotes viability, migration, and angiogenesis in human microvascular endothelial cells by sponging microRNA-195. J Cell Biochem. 2019;120(9):14360–14371. doi:10.1002/jcb.28692
31. Gao Z, Zhou H, Wang Y, Chen J, Ou Y. Regulatory effects of lncRNA ATB targeting miR-200c on proliferation and apoptosis of colorectal cancer cells. J Cell Biochem. 2019;121:332–343.
32. Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13:33. doi:10.1186/1476-4598-13-33
33. Ple H, Landry P, Benham A, et al. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7(12):e50746. doi:10.1371/journal.pone.0050746
34. Aleckovic M, Wei Y, LeRoy G, et al. Identification of Nidogen 1 as a lung metastasis protein through secretome analysis. Genes Dev. 2017;31(14):1439–1455. doi:10.1101/gad.301937.117
35. Pedrola N, Devis L, Llaurado M, et al. Nidogen 1 and nuclear protein 1: novel targets of ETV5 transcription factor involved in endometrial cancer invasion. Clin Exp Metastasis. 2015;32(5):467–478. doi:10.1007/s10585-015-9720-7
36. Rokavec M, Bouznad N, Hermeking H. Paracrine induction of epithelial-mesenchymal transition between colorectal cancer cells and its suppression by a p53/miR-192/215/NID1 axis. Cell Mol Gastroenterol Hepatol. 2019;7(4):783–802. doi:10.1016/j.jcmgh.2019.02.003
37. Li H, Zeng J, Shen K. PI3K/AKT/mTOR signaling pathway as a therapeutic target for ovarian cancer. Arch Gynecol Obstet. 2014;290(6):1067–1078. doi:10.1007/s00404-014-3377-3
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.