Back to Journals » OncoTargets and Therapy » Volume 11
LncRNA ANCR promotes proliferation and radiation resistance of nasopharyngeal carcinoma by inhibiting PTEN expression
Authors Ma X, Zhou J , Liu J, Wu G, Yu Y, Zhu H, Liu J
Received 3 August 2018
Accepted for publication 14 October 2018
Published 27 November 2018 Volume 2018:11 Pages 8399—8408
DOI https://doi.org/10.2147/OTT.S182573
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Xingkai Ma,1,* Jieyu Zhou,2,* Jianyong Liu,1 Geping Wu,1 Yan Yu,1 Hongyan Zhu,1 Jisheng Liu3
1Department of Otorhinolaryngology, Zhangjiagang First People’s Hospital, Affiliated Hospital of Soochow University, Suzhou, Jiangsu Province, P.R. China; 2Department of Otorhinolaryngology, Shanghai Ninth People’s Hospital, Shanghai Jiaotong University, School of Medicine, Shanghai, P.R. China; 3Department of Otorhinolaryngology, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu Province, P.R. China
*These authors contributed equally to this work
Introduction: Antidifferentiation noncoding RNA (ANCR) is a newly identified long noncoding RNA, which is reported to function as an oncogene in multiple human cancers. However, its function in nasopharyngeal carcinoma (NPC) and underlying mechanism are still unclear.
Materials and methods: We explored the expression of ANCR in NPC tissues and cells by real-time PCR and analyzed the relationship between ANCR expression and clinicopathological characteristics of NPC patients by Pearson’s chi-squared test. Then we inhibited ANCR expression in NPC cells using siRNAs and evaluated the effect of ANCR expression on cell proliferation, colony formation, and radiosensitivity by cell counting kit-8 assay and colony formation assay. We used RT-PCR and Western blot analyses to search target genes of ANCR. Also, we used RNA immunoprecipitation (RIP) assay and chromatin immunoprecipitation assay to study the molecular mechanism in this regulation.
Results: We found that ANCR was upregulated in NPC tissues and cells. ANCR expression was significantly correlated with tumor size and TNM stage. Further, ANCR knockdown inhibited NPC cell growth and radiation resistance. Mechanistically, we found that PTEN was upregulated in ANCR knockdown NPC cells. In addition, RIP assay indicated that EZH2, the oncogenic histone methyltransferase of polycomb repressive complex 2, interacted with ANCR in NPC cells. More importantly, the binding of EZH2 and deposition of relevant negative histone marker H3K27me3 on PTEN promoter depended on ANCR expression.
Conclusion: ANCR expression is upregulated in NPC and promotes NPC growth and radiation resistance through an epigenetic regulation of PTEN expression.
Keywords: antidifferentiation noncoding RNA, nasopharyngeal carcinoma, phosphatase and tensin homolog, growth, radiation resistance
Introduction
Nasopharyngeal carcinoma (NPC), originated from the mucosal epithelial cells of the nasopharynx, causes 50,000 deaths each year in the world and most of these patients are from Southern China and Southeast Asia. The incidence of NPC is considered to be associated with Epstein–Barr virus infection, hereditary and environmental factors.1,2 Currently, radiotherapy is the main therapeutic method for NPC patients.3 However, the 5-year survival rate of patients is still not ideal as the radiation resistance is a severe problem in treatment of advanced NPC.3 Thus, it is of great significance to explore the molecular mechanisms in NPC progression and radiation resistance, which may provide novel therapeutic targets for NPC.
Long noncoding RNAs (lncRNAs) are defined as a type of nonprotein coding transcripts with over 200 nucleotides. Recently, thousands of lncRNAs have been identified, and numerous studies have demonstrated that lncRNAs play vital roles in physiological and pathological processes.4–6 Antidifferentiation noncoding RNA (ANCR) was first identified as an lncRNA, which was required for suppression of progenitor cell differentiation.7 After that, several studies have revealed that ANCR was involved in tumorigenesis and progression. For instance, ANCR can increase the stemness features of hepatocellular carcinoma cells by regulating β-catenin expression.8 Moreover, ANCR is reported to promote migration and invasion of gastric cancer cells.9 In addition, the increased expression of ANCR is found to be associated with advanced tumor progression and poor prognosis in colorectal cancer patients.10 However, the biological function and significance of ANCR in NPC have not been reported.
In this study, we explored the potential role of ANCR in NPC progression. We found that ANCR expression was remarkably elevated in NPC tissues and cell lines. Moreover, the proliferation rate and resistance to radiation of NPC cells were suppressed when ANCR expression was knocked down. In addition, our result suggested that ANCR may repress phosphatase and tensin homolog (PTEN) expression synergistically with enhancer of zeste homolog 2 (EZH2), which is responsible for trimethylation of histone H3 lysine 27 (H3K27me3). Our study revealed the biological function and mechanism of ANCR in NPC progression and characterized ANCR as a potential biomarker and therapeutic target for NPC.
Materials and methods
Tissue sample collection
A total of 30 samples of NPC tissues and corresponding normal tissues were obtained from patients after complete or partial surgical resection at Zhangjiagang First People’s Hospital, during 2015–2017. No preoperative chemotherapy or radiotherapy treatment had been conducted in these patients before the operation. All specimens were immediately snap frozen in liquid nitrogen, and stored at −80°C until RNA extraction. The study was approved by the Scientific and Ethical Committee of Zhangjiagang First People’s Hospital and Institute, P.R. China. Written informed consent was obtained from all patients, and this procedure was conducted in accordance with the Declaration of Helsinki.
Cell culture
The human NPC cell lines (CNE1, CNE2, SUNE1 C666-1, HONE1, and HNE1) and human normal nasopharyngeal epithelial NP-69 cells were obtained from Shanghai Institutes for Biological Sciences Cell Resource Center. The cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (Invitrogen) and 1% (v/v) penicillin–streptomycin. All the cells were incubated at 37°C in a humidified air atmosphere containing 5% CO2.
Vector construction and cell transfection
For overexpressing EZH2, the full-length sequence with a FLAG tag at the N terminal was cloned into plasmid pcDNA3.1+ at BamH I and Xho I sites. The primers were 5′-CGGGATCCGACTACAAGGACGACGACGACAAGATGGGCCAGACTGGGAAGAA-3′ (forward) and 5′-CCCTCGAGTCAAGGGATTTCCATTTCTC-3′ (reverse). The siRNAs of ANCR were shown as below: 5′-GCCATTGAAGCTGGAATGT-3′ and 5′-GGCCAAATATGCGTACTAA-3′ (RiboBio Co. Ltd, Guangzhou, P.R. China). The negative control (NC) siRNA (5′-TTCTCCGAACGTGTCACGT-3′) was used as a negative control. All small molecules were transfected into cells by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection efficiency was measured 48 hours later.
Cell proliferation assay and colony formation assay
The cell proliferation rate was measured via the cell counting kit-8 (CCK-8) assay. NPC cells transfected with ANCR siRNAs or NC siRNA were seeded into 96-well plates (3,000 cells/well), cultured at daily intervals for 5 days, and then incubated with CCK-8 at 37°C for 2 hours. Optical density was determined at a wavelength of 450 nm using EnVision (PerkinElmer Inc., Waltham, MA, USA). After a 2-week incubation with 1,000 cells/well in 6-well plates, colony formation was observed by staining cells with 0.1% crystal violet (Sangon Biotech Co. Ltd, Shanghai, P.R. China).
Tumor xenografts in nude mice
All animal care and handling procedures were performed in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Zhangjiagang First People’s Hospital, P.R. China. For these studies, 6–8 week old female nude mice were purchased from the Model Animal Research Center of Nanjing University. 5×106 SUNE-1 cells in 100 μL of PBS: matrigel (9:1, v/v) were injected subcutaneously into each flank of nude mice (five mice per group). Tumor growth was evaluated every 4 days. The tumor volume was determined by measuring maximum (L) and minimum (W) length of the tumor using a slide caliper and calculated as ½LW2. Mice were euthanized at day 28, and tumors were collected, weighed, and analyzed.
RNA isolation and quantitative RT-PCR
Total RNA from tissues and cells was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). cDNAs were synthesized with a HiScript first Strand cDNA Synthesis kit (VAzyme Biotech, Nanjing, P.R. China). The FastStart Universal SYBR Green Master (Vazyme Biotech) was used for quantitative real-time PCR (qRT-PCR) according to the manufacturer’s instructions to measure the expression level of certain genes. Primers: p16, 5′-ATGGAGCCTTCGGCTGACT-3′ (forward), 5′-GTAACTATTCGGTGCGTTGGG-3′ (reverse), p21, 5′-TGTCCGTCAGAACCCATGC-3′ (forward), 5′-AAAGTCGAAGTTCCATCGCTC-3′ (reverse), p27, 5′-TAATTGGGGCTCCGGCTAACT-3′ (forward), 5′-TGCAGGTCGCTTCCTTATTCC-3′ (reverse), PTEN, 5′-TTTGAAGACCATAACCCACCAC-3′ (forward), 5′-ATTACACCAGTTCGTCCCTTTC-3′ (reverse), p57, 5′-GCGGCGATCAAGAAGCTGT-3′ (forward), 5′-GCTTGGCGAAGAAATCGGAGA-3′ (reverse), p53 5′-GAGGTTGGCTCTGACTGTACC-3′ (forward), 5′-TCCGTCCCAGTAGATTACCAC-3′ (reverse).
Western blot assay
Proteins were extracted from cells using RIPA buffer (Solarbio Science & Technology Co. Ltd., Beijing, P.R. China). Protein samples were separated through SDS-PAGE and transferred onto 0.22 μm PVDF membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked for 1 hour with 5% skim milk at room temperature. The primary antibody was incubated over night at 4°C, followed by incubation with a horseradish peroxidase-conjugated secondary antibody for 1 hour at room temperature. Immunoreactivity was visualized with enhanced chemiluminescence-chemiluminescent kit (Thermo Fisher Scientific, Waltham, MA, USA). Antibodies used were: PTEN (Abcam, Cambridge, UK, ab32199), GAPDH (MBL, Japan, M171-3), FLAG (Sigma, USA, F3156), EZH2 (Abcam, ab191250), and H3K27me3 (Abcam, ab6002).
RNA immunoprecipitation (RIP) assay
RIP assay was performed using the EZ-Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore). A total of 2×107 cells were harvested and lysed in 100 μL lysis buffer requiring for one RIP reaction. Five micrograms of FLAG antibody (Sigma, USA) and corresponding IgG were added into cell lysate and incubated with rotation overnight at 4°C. The immunoprecipitated RNA was purified using TRIzol regent and analyzed with qRT-PCR.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed with SUNE-1 cells using SimpleChIP® Plus Sonication Chromatin IP kit (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer’s instructions. Normal rabbit IgG served as the control. ChIP samples were analyzed by qRT-PCR using the FastStart Universal SYBR Green Master (Vazyme Biotech). The percentage of ChIP DNA was calculated relative to the input DNA from primer-specific standard curves. The primer sequences of PTEN promoter for ChIP are 5′-GGAGGCAGCCGTTCGGAGGATTATT-3′ (forward), 5′-GGAAATGGCTCTGGACTTGGCGGTA-3′ (reverse) and 3′UTR region primer sequences are 5′-TCCTGGATGACCTTTGACATAC-3′ (forward), 5′-TCCAGTATGCCAACTTTGGTT-3′ (reverse). Antibodies used were: EZH2 (Abcam, ab191250), H3K27me3 (Abcam, ab6002), RNA polymerase II (RNA polII) (Millipore, 05-623), and H3K4me3 (Abcam, ab8580).
Statistical analysis
All results were expressed as mean±SD from multiple independent experiments. The Student’s t-test was used to derive the significance of the differences between mean values. Data analysis was performed with the statistical program GraphPad Prism. A value of P<0.05 was considered to indicate a statistically significant result.
Results
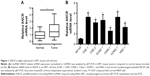
ANCR is highly expressed in NPC tissues and cell lines
To explore the biological function of lncRNA ANCR in NPC, we first examined the mRNA level of ANCR in NPC tissues and corresponding adjacent normal tissues from 30 patients using qRT-PCR. The expression of ANCR was significantly upregulated in NPC tissues (Figure 1A). We then analyzed the relationship between ANCR expression and the clinicopathological features. Notably, ANCR expression correlated with tumor size and TNM stage (Table 1). Further, we also examined ANCR expression in normal nasopharyngeal epithelial NP-69 cells and NPC cells, and consistently we found that ANCR was upregulated in six NPC cell lines, including CNE-1, CNE-2, SUNE-1, C666-1, HONE-1, and HNE-1 cells, compared with NP-69 cells (Figure 1B). Taken together, our data indicate that ANCR expression was upregulated in NPC tissues and cell lines and was associated with poor prognosis.
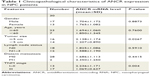  | Table 1 Clinicopathological characteristics of ANCR expression in NPC patients |
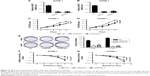
ANCR knockdown inhibits NPC cell growth and radiation resistance
To determine the effect of ANCR in NPC progression, we knocked down ANCR expression in NPC cell lines SUNE-1 and HONE-1 using small interference RNAs targeting ANCR (siANCR1 and siANCR2). qRT-PCR results indicated that ANCR expressions were significantly reduced in these cells transfected with siANCR compared with NC control cells (Figure 2A and B). We found that ANCR knockdown significantly impaired the growth of SUNE-1 and HONE-1 cells (Figure 2C and D). Additionally, colony formation assay suggested that ANCR knockdown led to a significant reduction in colony numbers compared with NC control cells (Figure 2E). To examine whether ANCRs have an impact on radiosensitivity, we exposed SUNE-1 and HONE-1 cells transfected with NC or siANCRs to the indicated doses of irradiation (0, 2, 4, 6, 8 Gy). The results suggested that ANCR knockdown enhanced radiosensitivity of NPC cells (Figure 2F and G). Thus, these results indicate that ANCR is crucial for NPC cell growth and radiosensitivity.
ANCR knockdown inhibits NPC cell proliferation in vivo
A tumor xenograft model was used to assess the effect of ANCR on tumor growth in vivo. ANCR knockdown SUNE-1 cells and NC SUNE-1 cells were subcutaneously injected into the different flanks of nude mice for 4 weeks. As shown in Figure 3, a marked reduction in tumor volume and weight in the siANCR1 groups was observed compared with that of NC group. These data indicate that ANCR promotes NPC cell proliferation in vivo.
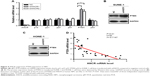
ANCR suppresses PTEN expression in NPC
We next investigated potential mechanisms by which ANCR knockdown impaired proliferation and enhanced radiosensitivity of NPC cells. We analyzed the expression of a series of proliferation-related genes, including p16, p21, p27, p57, PTEN, and p53 in SUNE-1 cells by qRT-PCR. We found that the expression of PTEN, a famous tumor suppressor, was greatly increased in ANCR knockdown SUNE-1 cells compared with NC control cell (Figure 4A). Then we preformed Western blot analysis to confirm this effect of ANCR on PTEN expression at the protein level. The results suggested that PTEN expression levels were increased in ANCR knockdown cells (Figure 4B and C). Further, qRT-PCR data in NPC tissues exhibited a significant inverse correlation between ANCR and PTEN mRNA levels (Figure 4D). These results suggest that ANCR suppresses PTEN expression in NPC cells.
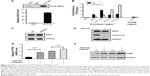
ANCR-mediated PTEN silencing depends on EZH2
LncRNA can function as molecular scaffold to help epigenetic enzymes to bind on promoters of target genes.5 It was reported that ANCR interacted with enhancer of EZH2, a core component of polycomb repressive complex 2 (PRC2), which catalyzes trimethylation of H3K27me3 and leads to subsequent gene silencing.11,12 To investigate their interaction in NPC cells, we did RIP assay in FLAG-tagged EZH2 overexpressed SUNE-1 cells. As shown in Figure 5A, FLAG–EZH2 was detected in the immunoprecipitates using Western blot assay, and ANCR was also detected by qRT-PCR. Then, to explore the potential mechanism of ANCR and EZH2 in PTEN regulation, we did (ChIP) assay using SUNE-1 cells transfected with NC or siANCR1. The result suggested that ANCR knockdown resulted in decreased EZH2 binding and H3K27me3 enrichment on PTEN promoter. In addition, we found that RNA polII binding and active histone marker H3K4me3 level was increased on PTEN promoter in ANCR knockdown cells. This result was consistent with activated PTEN expression (Figure 5B). Thus, it is believed that ANCR is required for EZH2 binding on PTEN promoter. It was reported that ANCR can regulate EZH2 expression and degradation.13,14 To examine this possibility in NPC cells, we used Western blot to check the protein level of EZH2 and the result showed that ANCR did not affect EZH2 expression in NPC cells (Figure 5C). These data indicated that EZH2 binding on PTEN promoter was ANCR dependent. Then, we used GSK126, an inhibitor of EZH2 catalytic activity, to treat with SUNE-1 cells. Western blot results showed that histone marker H3K27me3 was significantly decreased after 10 μm GSK126 treating for 48 hours in SUNE-1 cells, without change in the protein level of EZH2 (Figure 5D). We next examined PTEN mRNA and protein level in SUNE-1 cells treated with GSK126 and ANCR siRNA separately or together. As shown in Figure 5E and F, PTEN mRNA and protein level was increased after GSK126 treating or ANCR knockdown. Intriguingly, ANCR knockdown led to less fold changes of PTEN mRNA levels in GSK126-treated SUNE-1 cells (3.52 vs 1.26, Figure 5E). PTEN protein-level changes were similar with mRNA-level changes, which indicated that ANCR-mediated PTEN repression is EZH2 catalytic activity dependent (Figure 5F). Taken together, these results suggest that EZH2 and ANCR synergistically regulate PTEN expression and EZH2 is necessary for ANCR to repress the transcription of PTEN in SUNE-1 cells.
Discussion
Accumulating evidence has revealed that lncRNAs play crucial roles in human diseases, such as cancers, cardiovascular diseases, diabetes, and the like.15–17 Numerous studies have reported the relationship between lncRNAs and different aspects of tumorigenesis, such as proliferation, migration, invasion, metastasis, resistance to chemotherapy and radiotherapy, and so on.18–20 LncRNA ANCR is a newly identified noncoding RNA that is found to be overexpressed in multiple types of cancers and acts as an oncogene to promote cancer progression.8–10,21–23 In this study, we found that ANCR expression was significantly elevated in NPC tissues and cell lines and was well associated with tumor size and TNM stage. Previous studies have demonstrated that certain lncRNAs may reflect the progress of cancers and can be regarded as potential biomarkers for clinical diagnosis.24,25 Our data suggested that ANCR may be a candidate predictor for early diagnosis and clinical treatment for NPC patients.
Radiotherapy is the mainstay treatment for NPC patients and has been shown to improve patients’ survival. However, radiation resistance is still an obstacle in treatment for advanced NPC. PTEN, a well-studied tumor suppressor gene, can regulate the PI3K/AKT pathway by catalyzing the dephosphorylation of phosphatidylinositol 3-, 4-, 5-trisphosphate.26 It was reported that PTEN/PI3K/AKT pathway was involved in transformation of cancer stem-like cells (CSCs), which were shown to be resistant to radiation.27–29 Zhang and his colleagues have demonstrated that NPC cells without PTEN expression have CSCs properties and are resistant to radiation.30 Here, we report that lncRNA ANCR can promote NPC cell growth and radiation resistance, which was mainly dependent on PTEN.
EZH2, which is responsible for H3K27me3 formation and function as an epigenetic suppressor, is found to be overexpressed in multiple cancers and acts as an oncogene to promote tumor progression.31–33 In this study, we found that ANCR knockdown resulted in reduced binding of EZH2 on PTEN promoter, which suggested that ANCR may play a role in binding of EZH2 on promoters of downstream target genes. EZH2 is the functional enzymatic component of PRC2 complex, which contains other proteins such as SUZ12, JARID2, AEBP2, EED, RbAp46/48, and polycomblike.34 Of note, epigenetic enzymes such as DNA methyltransferases, histone deacetylases, and Sirt1 can also transiently interact with PRC2 complex.34 Our data indicate that ANCR may act as a scaffold to stable the interaction of EZH2 and other proteins within or without PRC2 complex, and this effect is vital in the regulation of EZH2 target genes.
Conclusion
Our results demonstrate that lncRNA ANCR promotes NPC cell growth and radiation resistance by repressing the expression of PTEN. This regulation relies on ANCR-mediated EZH2 binding and epigenetic regulation on PTEN promoter. Our data suggest that ANCR may represent as a potential biomarker and therapeutic target for radiation resistance in NPC patients.
Acknowledgments
This work was supported by the Youth Science and Technology Project of Suzhou (KJXW2016053), grants from Bureau of Science and Technology of Zhangjiagang (ZKS1516) and Jiangsu University (JLY20160112).
Disclosure
The authors report no conflicts of interest in this work.
References
Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46(8):866–871. | ||
Petersson F. Nasopharyngeal carcinoma: a review. Semin Diagn Pathol. 2015;32(1):54–73. | ||
Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012–1024. | ||
Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics. 2017;17(2–3):135–143. | ||
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. | ||
Mallardo M, Poltronieri P, D’Urso OF. Non-protein coding RNA biomarkers and differential expression in cancers: a review. J Exp Clin Cancer Res. 2008;27:19. | ||
Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26(4):338–343. | ||
Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63(2):499–511. | ||
Mao Z, Li H, du B, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37(6):BSR20171070. | ||
Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11480–11484. | ||
Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7(3):299–313. | ||
Zhu L, Xu PC, Pc X. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432(4):612–617. | ||
Yang ZY, Yang F, Zhang YL, et al. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark. 2017;18(1):95–104. | ||
Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24(1):59–71. | ||
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. | ||
Chen G, Wang Z, Wang D, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41(Database issue):D983–D986. | ||
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. | ||
Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. | ||
Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152–166. | ||
Chen QN, Wei CC, Wang ZX, Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2017;8(1):1925–1936. | ||
Jia J, Li F, Tang XS, et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7(25):37868–37881. | ||
Lu QC, Rui ZH, Guo ZL, Xie W, Shan S, Ren T. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J Cell Mol Med. 2018;22(3):1527–1537. | ||
Sha S, Yuan D, Liu Y, Han B, Zhong N. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol Open. 2017;6(9):1310–1316. | ||
Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. | ||
Zhang W, Wang L, Zheng F, et al. Long noncoding RNA expression signatures of metastatic nasopharyngeal carcinoma and their prognostic value. Biomed Res Int. 2015;2015:618924. | ||
Georgescu MM. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer. 2010;1(12):1170–1177. | ||
Li B, Lu Y, Wang H, et al. miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother. 2016;79:93–101. | ||
Li H, Gao Q, Guo L, Lu SH, Hx L, Sh L. The PTEN/PI3K/Akt pathway regulates stem-like cells in primary esophageal carcinoma cells. Cancer Biol Ther. 2011;11(11):950–958. | ||
Dubrovska A, Kim S, Salamone RJ, et al. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc Natl Acad Sci U S A. 2009;106(1):268–273. | ||
Zhang G, Wang W, Yao C, et al. Radiation-resistant cancer stem-like cell properties are regulated by PTEN through the activity of nuclear β-catenin in nasopharyngeal carcinoma. Oncotarget. 2017;8(43):74661–74672. | ||
Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. | ||
Sellers WR, Loda M. The EZH2 polycomb transcriptional repressor – a marker or mover of metastatic prostate cancer? Cancer Cell. 2002;2(5):349–350. | ||
Melling N, Thomsen E, Tsourlakis MC, et al. Overexpression of enhancer of zeste homolog 2 (EZH2) characterizes an aggressive subset of prostate cancers and predicts patient prognosis independently from pre- and postoperatively assessed clinicopathological parameters. Carcinogenesis. 2015;36(11):1333–1340. | ||
Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.