Back to Journals » Cancer Management and Research » Volume 13
lncRNA AC007207.2 Promotes Malignant Properties of Osteosarcoma via the miR-1306-5p/SIRT7 Axis
Authors Dang Y, Zhou Y, Ou X, Wang Q, Wei D, Xie F
Received 7 May 2021
Accepted for publication 8 September 2021
Published 21 September 2021 Volume 2021:13 Pages 7277—7288
DOI https://doi.org/10.2147/CMAR.S318975
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Youting Dang,1 Yunping Zhou,2 Xuehai Ou,2 Qiang Wang,2 Dengke Wei,2 Fei Xie2
1Department of Pediatric Orthopedics, Honghui Hospital, Xi’an Jiao Tong University, Xi’an, Shaanxi, 710054, People’s Republic of China; 2Department of Hand Surgery, Honghui Hospital, Xi’an Jiao Tong University, Xi’an, Shaanxi, 710054, People’s Republic of China
Correspondence: Fei Xie
Department of Hand Surgery, Honghui Hospital, Xi’an Jiao Tong University, 555 East Friendship Road, Xi’an, Shaanxi, 710054, People’s Republic of China
Email [email protected]
Background: Long non-coding RNAs (lncRNAs) have been implicated in initiation and development of numerous cancers. In the present study, we explored the role of lncRNAs AC007207.2 in osteosarcoma (OS).
Methods: Gene expression data of OS tissues was downloaded from the TARGET database. All the experiments were repeated at least three times. Data were analyzed using Perl, R, SPSS v12.0 and GraphPad Prism 8 software.
Results: We found lncRNA AC007207.2 was over-expressed in OS tissues and cell lines, and this phenomenon was associated with the worse prognosis of OS. Moreover, we found that AC007207.2 promotes proliferation and metastasis of OS cells via the miR-1306-5p/SIRT7 axis. Meanwhile, we found miR-1306-5p remarkably inhibits the malignant behavior of OS cells.
Conclusion: lncRNA AC007207.2 promotes progression of OS by upregulating SIRT7 expression through miR-1306-5p sponging. Thus, lncRNA AC007207.2/miR-1306-5p/SIRT7 axis is a promising therapeutic target for OS treatment.
Keywords: LncRNA AC007207.2, osteosarcoma, miRNA
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor, predominantly affecting children and adolescents. In particular, it is the eighth leading cause of childhood cancer, with its incidence rate reaching 2.4%.1 Currently, neoadjuvant chemotherapy combined with surgery are the first-line treatment options for OS. The overall five-year survival rate of OS is around 68%.2 Unfortunately, pulmonary metastasis in the early stage worsens the OS prognosis. Acidity, hypoxia, and chemokines in the bone microenvironment have all been linked with metastasis and progression of OS.3 OS patients respond differently to recent advanced treatments such as PDT, molecular targeted immunotherapy, and genetic engineering based modalities.4
Competing endogenous RNAs (ceRNAs) regulate other RNA transcripts by competing for shared microRNAs (miRNAs).5 MiRNAs are non-coding transcripts that regulate gene expression post-transcriptionally.6 To some extent, abnormal ceRNA expression is a biomarker for progression of certain tumors. Wang et al reported that overexpression of TUG1 is associated with poor prognosis of OS.7 Chen et al further reported that MIR4435-2HG and ELFN1-AS1, all components of ceRNA network, are prognostic markers of colorectal carcinoma.8 Also, Han et al found that the stemness of lung adenocarcinoma was regulated via the ceRNA axis.9 Overall, the relationship between multiple members of ceRNA system and cancers remain to be elucidated.
In this study, we explored the role of lncRNA AC007207.2 in OS patients. qRT-PCR revealed that lncRNA AC007207.2 was over-expressed in OS tissue and cell lines. Meanwhile, over-expression of AC007207.2 was associated with shorter overall survival of OS patients. In vitro experiments further showed that AC007207.2 knockdown significantly modulated the proliferation and invasion of OS cells. Further analyses revealed that AC007207.2 promotes its pro-cancer functions via the miR-1306-5p/SIRT7 axis.
Materials and Methods
Bioinformatics Analysis
Transcriptome data and clinical information of OS patients were downloaded from the Therapeutically Applicable Research To Generate Effective Treatments (TARGET) database. Prior to the analysis, all the samples were preprocessed with the following steps: 1. probes annotation; 2. combination of expression profile and clinical features; 3. detection of low abundance gene; 4. missing value complement. LncRNAs were distinguished from mRNAs using biotype data in the “Homo_sapiens.GRCh37.gtf” file. Differentially expressed lncRNAs (DELs) between OS and matched normal tissues were identified using the “Limma” package in R software. Statistical significance in expression of DELs was set at |logFC>1| and P < 0.05. The relationship between lncRNAs AC007207.2 expression and prognosis of OS patients was analyzed using Kaplan–Meier survival curves.
Tissues and Cell Lines
OS and matched tissues analyzed in this study were all extracted from patients attending the Honghui Hospital Affiliated to Medical School of Xi’an Jiao Tong University. A total of 24 osteosarcoma and 13 normal tissues were extracted. All patients consented to participate in this research. The protocol for this study was approved by Research Ethics Committee of Honghui Hospital. This study was performed according to the Helsinki Declaration. Four osteosarcoma cell lines (U-2OS, Saos-2, 143B, MG63) and one osteoblast cell line (hFOB1.19) were purchased from the cell library of the Chinese Academy of Sciences.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA from tissues and OS cell lines was extracted using Trizol (ThermoFisher, USA). The RNA was then reversed transcribed to cDNA using the cDNA reverse transcription kit (Invitrogen) and thereafter amplified using qPCR. The sequence of primers used in the for PCR process are as follows: AC007207.2, forward primer: 5ʹ-CCTATTCACCCAGGACCACA-3ʹ, reverse primer: 5ʹ-TGGCAAACATCAACTCCCTC-3ʹ; SIRT7, forward primer: 5ʹ-GACCTGGTAACGGAGCTGC-3ʹ, reverse primer: 5ʹ-CGACCAAGTATTTGGCGTTCC-3ʹ; miR-1306-5p, reverse transcription primer: 5ʹ- GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGACG-3ʹ, forward primer: 5ʹ- AGACCACCTCCCCTGCAAAC-3ʹ, reverse primer: 5ʹ- CAGTGCAGGGTCCGAGGT-3ʹ.
Cell Transfections
AC007207.2 knockdown was performed using shRNAs (sh-AC007207.2). The sequence of shRNAs used in this study were 5ʹ-CAGATTCACTATAACTGTCATTCCA-3ʹ for shRNA1, 5ʹ-CACATCAAGCAGAGGAGAAAGATAA-3ʹ for shRNA2, and 5ʹ- GGATCATAATGGATGTTGT-3ʹ for shRNA3. The sequence of shRNA for SIRT7 (sh-SIRT7) was 5ʹ-CATTGAAGTCTGTACCTCCTGCGTT-3ʹ. The miR-1306-5p mimics and inhibitor were purchased from RiboBio (Guangzhou, China). Transfection was performed using the Lipofectamine 2000 transfection kit (Invitrogen). The cells transfected with scrambled plasmids were defined as control group (sh-NC, NC mimics and NC inhibitor).
Western Blotting
Total proteins were extracted using a total protein extraction kit (KeyGen Biotech, Nanjing, China) when cells were cultured to 90–100% confluency. Then, protein samples were mixed with loading buffer and then denatured by heating 10 min. The samples were resolved in a SDS/PAGE and then transferred into membrane. The membrane was incubated with primary antibodies (Proteintech: anti-Bax, 1:1000; anti-Bcl-2, 1:1000, anti-Cleaved-caspase-3, 1:1000, anti-GAPDH, 1:5000) overnight at 4°C. The blots were washed in 1X PBS and then incubated with a secondary antibody for 2 h at room temperature.
Luciferase Reporter Assay
The correlation between AC007207.2 expression compared with both SIRT7 and miR-1306-5p expression were analyzed using the luciferase reporter assay. psiCHECK-2-AC007207.2-wild-type (AC007207.2-wt), psiCHECK-2-AC007207.2-mutant (AC007207.2-mut), psiCHECK-2-SIRT7-wild-type (SIRT7-wt) and psiCHECK-2-SIRT7-mutant (SIRT7-mut) vectors with binding sites were purchased from GenePharma (Shanghai, China). In brief, the mutant sequences were created using PCR methods and cloned into reported plasmids. The dual-luciferase plasmids were created in the psiCheck2 vector (Promega), with firefly luciferase serving as a control, and the various Gars target regions cloned downstream of the Renilla luciferase stop codon. The miR-1306-5p mimics and control plasmids were purchased from RiboBio (Guangzhou, China). Transfection was performed using Lipofectamine 2000. Reporter vectors were co-transfected with miR-1306-5p mimics or inhibitor. Luciferase activity was assessed after 48 hours of transfection using the Dual-Luciferase Reporter Assay System (Promega).
Colony Formation Assay
This assay was used to assess the effect of AC007207.2 expression on proliferation of cancer cells. Briefly, cultured cells were centrifuged and resuspended in serum-free conditioned medium then seeded into six-well plates at the rate of 500 cells per well. The culture medium was changed every five days. Cells were fixed and stained using crystal violet.
CCK8 Assay
CCK8 assay was performed to evaluate the effect of AC007207.2 expression on viability of OS cells. The experiment was performed using a CCK8 kit (Dojindo, Shanghai). The (143B and MG63) cell lines were collected and resuspended in serum-free conditioned medium and thereafter plated into a 96-well plate at the rate of 1×104 cells per well. CCK8 reagents were then added to the plates before culturing the cells for 2 hours at 37°C. The proliferation rate of cells was based on OD value, measured at 450 nm.
Cell Cycle and Apoptosis Assay
The effect of lncRNAs AC007207.2 on cycle and apoptosis of 143B and MG63 cells were analyzed using flow cytometry. For the cell cycle analysis, cells were digested and resuspended overnight in 75% ice-cold ethanol and thereafter stained for 30 min in the dark using Propidium Iodide (PI)/RNase solution. For apoptosis analysis, cells were digested and resuspended in a binding buffer. Dead cells were stained using Annexin V-FITC system before flow cytometric analysis.
Transwell Assay
Transwell assay including cell invasion and migration was performed in 24-well plates. The upper chamber contained Matrigel, whereas the lower chamber contained 800ul of serum-free conditioned medium supplemented with 10% serum. Cells were then seeded into the upper chamber for 12 hours, fixed with anhydrous ethanol and stained using crystal violet.
Scratch-Healing Assay
The effect of lncRNAs AC007207.2 on migration of 143B and MG63 cell lines was analyzed using the scratch-healing assay. Cells were seeded into six-well plates and cultured to 80–90% confluence. A scratch was made in the cell monolayer using a 200ul tip. The cells were cultured in a serum-free medium for 24h. The scratches were then photographed at 0 and 24 hours under (an electron scanning microscope).
RNA Fluorescence in situ Hybridization (FISH)
The expression of lncRNAs AC007207.2 in cancer tissue and cell lines was assessed using FISH assay. OS tissues of patients with high and low AC007207.2 expression were frozen, cut into thin 5 μm sections fixed, washed, and blocked with PBS supplemented with 2% BSA and 5% FBS. Fluorescent labeled oligonucleotide probes complementary to AC007207.2 (5ʹ-ATATGGATCTCTTGTGGAGGAACC-3ʹ) were purchased from GenePharma (Shanghai, China). The subcellular localization of AC007207.2 was also identified by FISH using a FISH Kit (RiboBio, Guangzhou, China). Images were captured using a confocal microscope.
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
The effect of AC007207.2 on proliferation of OS cells and cell lines was assessed using EdU assay, based on the BeyoClick EdU-647 staining kit. Briefly, 5×105 cells were seeded into a six-well plate before adding EdU solution when the cells adhered to the wall. Nuclei were stained using DAPI solution. Fluorogenic signals were captured using an inverted fluorescence microscope.
RNA Pulldown Assay
Two dishes of (143B and MG63 cell lines) cells were digested and pulled together in one tube. Approximately 1×107 of the cells were then counted and synchronized for 3 min at 1000 rpm. The cells were then lysed for 30 min using ice col NP40 and spinning for 10 min at 12,000 rpm. Biotin-labeled AC007207.2 probe and miR-1306-5p probes and their corresponding antisense probes were designed and synthesized at GenePharma (Shanghai, China). Briefly, cell lysates were incubated for 2 hours at room temperature in a mixture of biotinylated probes and 40 μL of streptavidin agarose beads (Invitrogen Life Technologies, CA, USA). RNA pulldown from the biotinylated probes were performed using PCR.
RNA Immunoprecipitation (RIP) Assay
RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, CA, USA). Here, cell lysates were incubated for 30 min with anti Ago2 and IgG antibody on magnetic beads. The immunoprecipitated RNA was purified using Cold RIP wash buffer.
Statistical Analysis
Data were analyzed using SPSS, GraphPad Prism and R software. Two sided P < 0.05 was regarded statistically significant. All the experiments were performed in triplicate at least three times.
Results
LncRNA AC007207.2 is Over-Expressed in OS Tissue and Cell Lines
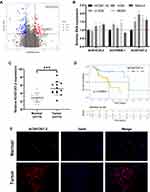
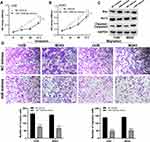
A brief flowchart of the entire study is shown in Figure S1. We identified 135 differentially expressed lncRNAs (DELs) between primary and metastatic OS tissue. Among these, lncRNA AC015726.2 (Normal mean = 0.008, Tumor mean = 0.525), AC078906.1 (Normal mean = 0.079, Tumor mean = 0.682), and AC007207.2 (Normal mean = 0.273, Tumor mean = 2.204) were the top three over-expressed lncRNAs (Figure 1A). Given that lncRNA AC007207.2 was stably overexpressed across the four OS cell lines (Figure 1B), it was used in our subsequent analyses. AC007207.2 was also over-expressed in OS tissue, relative to normal tissues (13 pairs of normal and OS tissues) (Figure 1C). Kaplan–Meier survival curve analysis demonstrated that over-expression of AC007207.2 was strongly associated with poor prognosis of OS patients (Figure 1D). FISH analyses further revealed that there were more AC007207.2 positive cells in tumor tissue, relative to normal tissue (Figure 1E).
LncRNA AC007207.2 Promotes Metastasis of OS Cells
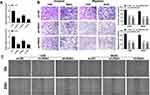
qRT-PCR demonstrated efficient knockdown of AC007207.2 by siRNA#1 and siRNA#3 in 143B and MG63 cell lines, respectively (approximate target efficiency, MG63 cells, shRNA1 = 65.9%, shRNA2 = 36.6%, shRNA3 = 70.4%; 143B cells, shRNA1 = 64.3%, shRNA2 = 38.8%, shRNA3 = 73.2%). Thus, siRNA#1 and siRNA#3 were used in the subsequent experiments (Figure 2A). Transwell assay revealed that AC007207.2 knockdown significantly reduced the invasion and migration of both 143B and MG63 cell lines, implying that AC007207.2 promotes metastasis of OS cells (Figure 2B). Scratch-healing assay revealed comparable findings, in which the scratch area for AC007207.2 knockdown group was wider than that of the control group (Figure 2C).
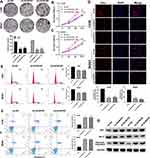
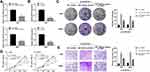
LncRNA AC007207.2 Promotes Proliferation of OS Cells
Colony formation assay revealed that AC007207.2 knockdown markedly inhibited clone formation ability of OS cells (Figure 3A), consistent with CCK8 assay analyses (Figure 3B and C). EdU assay demonstrated that inhibiting AC007207.2 expression significantly impaired proliferation of OS cancer cells (Figure 3D). Considering that apoptosis and cell cycle processes affects proliferation of cells, we further explored the effect of AC007207.2 on these processes. Flow cytometric analysis revealed that AC007207.2 knockdown disrupted the cells cycle, with only a small proportion of cells progressing to the S-phase (Figure 3E). Also, AC007207.2 knockdown promoted apoptosis of OS cells (Figure 3F). Further analyses revealed that AC007207.2 knockdown up-regulated the expression of Bax and cleaved-Cas3 proteins but modulated that of Bcl-3, all which participates in the proliferation of cells (Figure 3G).
AC007207.2 Promotes OS Cancer Properties by Binding miR-1306-5p
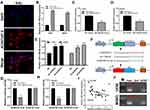
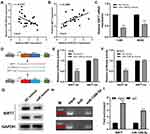
FISH assay revealed that AC007207.2 is mainly localized in the cytoplasm, implying that it function like ceRNA (Figure 4A). Bioinformatics analysis of miRNAs data in the Starbase database (http://starbase.sysu.edu.cn/index.php) revealed that miR-1306-5p was the most probable AC007207.2 target. Therefore, miR-1306-5p mimics were transfected into 143B and MG63 cells to explore interaction between this miRNA and AC007207.2. Transfection efficiency of miR-1306-5p in 143B and MG63 cell lines was validated using qRT-PCR (Figure 4B). We found that overexpression of miR-1306-5p significantly modulates the expression of AC007207.2 mRNA (Figure 4C and D). Interestingly, AC007207.2 knockdown substantially up-regulated the expression of miR-1306-5p (Figure 4E). Mutations were then induced in predicted binding sites of AC007207.2 on miR-1306-5p were then mutated (Complementary AC007207.2 sequences: GGAGGUG; Complementary miR-1306-5p sequences: CCUCCAC; AC007207.2 mutant sequences, AC007207.2: GGAGGUG to CCUCCAC) (Figure 4F). Dual-luciferase reporter gene assay revealed that miR-1306-5p mimics remarkably reduced the activity of AC007207.2 3′-UTR-reporter gene, relative to mutant AC007207.2 3′-UTR-reporter gene (Figure 4G and H). PCR revealed a strong negative correlation between AC007207.2 and miR-1306-5p expression (Figure 4I; r = −0.1702, P < 0.01). RNA pulldown assay demonstrated that the biotin-AC007207.2 probe could enrich more miR-1306-5p than the anti-probe (Figure 4J), indicating that AC012213.3 could bind to miR-1306-5p.
MiR-1306-5p Suppress the Progression of OS Cells and Expression of SIRT7
We further explored the biological role of miR-1306-5p in OS. CCK8 assay revealed that over-expression of miR-1306-5p inhibited proliferation of OS cells (Figure 5A and B). Western blot further revealed that overexpression of miR-1306-5p increased the expression of Bax and cleaved-Cas3, but modulated that of Bcl-2 (Figure 5C). Transwell assay further demonstrated that over-expression of miR-1306-5p significantly suppressed the invasion and migration of OS cells (Figure 5D). Given that miR-1306-5p targets SIRT7 gene, we speculated that it regulates progression of OS by binding SIRT7.10 Indeed, further analyses revealed a strong negative correlation between miR-1306-5p and SIRT7 expression in OS tissue (Figure 6A; r = - 0.3177; P < 0.01). Interestingly, we observed a positive correlation between AC007207.2 and SIRT7 expression (Figure 6B; r = 0.3366; P < 0.01). Meanwhile, overexpression of miR-1306-5p decreased the transcription of SIRT7 mRNA in OS cell lines (Figure 6C). Analysis of Starbase data identified the most probable miR-1306-5p binding sites on SIRT7 (Complementary SIRT7 sequences: UCCGUAGGAGGUG; Complementary miR-1306-5p sequences: ACGUCCCCUCCAC; SIRT7 mutant sequences: UCCGUAGGAGGUG to ACGUCCCCUCCAC) (Figure 6D). Dual-luciferase reporter gene experiment demonstrated that overexpression of miR-1306-5p inhibited the luciferase activities of wild type SIRT7, but had no noticeable effect on SIRT7 mutant (Figure 6E and F). Western blot showed that miR-1306-5p could significantly decrease SIRT7 protein expression (Figure 6G). RNA pulldown assay revealed that the biotin-miR-1306-5p probe could enrich more SIRT7 than the anti-probe (Figure 6H). RIP assay further demonstrated that Ago2 protein binds more to SIRT7 and miR-1306-5p than control lgG, implying that SIRT7 interacts with miR-1306-5p (Figure 6I).
LncRNA AC007207.2 Promotes Progression of OS via the miR-1306-5p/SIRT7 Axis
We further knocked down miR-1306-5p and SIRT7 in 143B and MG63 cells to explore the association between these miRNA and expression of SIRT7 (Figure 7A and B). CCK8 and colony formation assay revealed that miR-1306-5p inhibition induced proliferation of cells expressing sh-AC007207.2, whereas SIRT7 knockdown partially impaired this phenomenon (Figure 7C and D), consistent with transwell assay analyses. miR-1306-5p knockdown substantially reversed the inhibitory effect of downregulating AC007207.2 expression, similar to SIRT7 knockdown (Figure 7E). In general, our findings demonstrated that lncRNA AC007207.2 promotes proliferation and invasion of OS cells via the miR-1306-5p/SIRT7 axis.
Discussion
OS is the most prevalent bone cancer, and the second most common cancer in the elderly.11 Owing to its high malignancy and poor prognosis, the efficacy of OS treatment is still sub-optimal.1 This underlines the need to identify accurate biomarkers and efficient therapeutic targets for OS treatment.
To the best of our knowledge, this is the first study exploring the mechanism underlying the effect of AC007207.2 in OS tissues. We found that AC007207.2 is over-expressed in OS tissue and cell lines. Meanwhile, AC007207.2 promotes proliferation and invasion of OS cells in a manner similar to ceRNA (AC007207.2/miR-1306-5p/SIRT7).
In living organisms, particularly mammals, miRNAs regulate numerous patho and physiological functions.12 For example, miR-542-3p disrupts migration, invasion and EMT progress of HCC cells by targeting UBE3C.13 Chai et al reported that miR-19a-3p regulates ischemia/reperfusion induced inflammation and apoptosis by targeting IGFBP3.14 MiR-1306-5p regulates numerous biological processes under multiple diseases pathologies.10,15,16 For instance, Zheng et al revealed that miR-1306-5p regulates proliferation, migration and invasion of vascular smooth muscle cells by targeting lncRNA SNHG7-003.10 Elsewhere, Genis et al found that the expression of miR-1306-5p was strongly associated with prognosis of patients with cardiovascular complications.17 However, the role of miR-1306-5p in cancer tissues is not well understood. In the present study, we found lncRNA AC007207.2 promotes proliferation of OS cells via the miR-1306-5p/SIRT7 axis.
AC007207.2 performs its function in a manner similar to ceRNA (miR-1306-5p/SIRT7 axis). CeRNAs regulate numerous biological functions through a complex network.18 MiRNAs have been implicated in the development and progression of numerous cancers. For example, Zhang et al demonstrated that lncRNA SNHG8 promotes proliferation and invasion in gastric cancer cells via the miR-491/PDGFRA axis.19 In a related study, Meng et al revealed that lncRNA MIR99AHG promotes EMT of gastric cancer cells via the miR577/FOXP1 axis.20 Meanwhile, miR-151a-3p regulates the malignant behavior of OS by targeting lncRNA SNHG3.21 SIRT7 is a member of the sirtuin family and promotes proliferation and migration of tumor cells as well as development of metabolic disorders and multiple diseases.22 SIRT7 induces EMT and activates TGF-β signaling in breast cancer tissues by promoting degradation of SMAD4 via the β-TrCP1 pathway.23 SIRT7 enhance the activity of p53 (arresting the cell-cycle) by promoting the degradation of MDM2 via the PCAF signaling pathway.24 SIRT7 promotes the proliferation and invasion of OS cells by modulating binding of H3K18ac on the promoter region of CDC4 gene.25 Herein, we found that AC007207.2 promotes OS progression via the miR-1306-5p/SIRT7 axis. Nevertheless, more investigation on clinical level and more intensive molecular mechanisms are also needed to reinforce these findings.
Conclusion
Summarily, AC007207.2 is over expressed in OS tissues and this phenomenon is associated with worse prognosis of OS. In particular, AC007207.2 promotes proliferation and invasion of OS cells partly through the miR-1306-5p/SIRT7 axis. As such, AC007207.2 is a potential therapeutic targets in OS patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–vii325.
2. Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543.
3. Yang C, Tian Y, Zhao F, et al. Bone microenvironment and osteosarcoma metastasis. Int J Mol Sci. 2020;21:19.
4. Yu W, Wang Y, Zhu J, et al. Autophagy inhibitor enhance ZnPc/BSA nanoparticle induced photodynamic therapy by suppressing PD-L1 expression in osteosarcoma immunotherapy. Biomaterials. 2019;192:128–139.
5. Fiannaca A, Paglia L, Rosa M, Rizzo R, Urso A. miRTissue (ce): extending miRTissue web service with the analysis of ceRNA-ceRNA interactions. BMC Bioinform. 2020;21(Suppl 8):199.
6. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222.
7. Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108(5):859–867.
8. Chen J, Song Y, Li M, et al. Comprehensive analysis of ceRNA networks reveals prognostic lncRNAs related to immune infiltration in colorectal cancer. BMC Cancer. 2021;21(1):255.
9. Han P, Yang H, Li X, et al. Identification of a novel cancer stemness-associated ceRNA axis in lung adenocarcinoma via stemness indices analysis. Oncol Res. 2020;28:7.
10. Zheng J, Tan Q, Chen H, et al. lncRNA‑SNHG7‑003 inhibits the proliferation, migration and invasion of vascular smooth muscle cells by targeting the miR‑1306‑5p/SIRT7 signaling pathway. Int J Mol Med. 2021;47(2):741–750.
11. Corre I, Verrecchia F, Crenn V, Redini F, Trichet V. The osteosarcoma microenvironment: a complex but targetable ecosystem. Cells. 2020;9:4.
12. Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci. 2019;20:24.
13. Tao J, Liu Z, Wang Y, et al. MiR-542-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting UBE3C. Biomed Pharmacother/ Biomedecine Pharmacotherapie. 2017;93:420–428.
14. Chai Z, Gong J, Zheng P, Zheng J. Inhibition of miR-19a-3p decreases cerebral ischemia/reperfusion injury by targeting IGFBP3 in vivo and in vitro. Biol Res. 2020;53(1):17.
15. Chen X, Li C, Li J, Sheng L, Liu X. Upregulation of miR-1306-5p decreases cerebral ischemia/reperfusion injury in vitro by targeting BIK. Biosci Biotechnol Biochem. 2019;83(12):2230–2237.
16. Hindle AG, Thoonen R, Jasien JV, et al. Identification of candidate miRNA biomarkers for glaucoma. Invest Ophthalmol Vis Sci. 2019;60(1):134–146.
17. Bayés-Genis A, Lanfear DE, de Ronde MWJ, et al. Prognostic value of circulating microRNAs on heart failure-related morbidity and mortality in two large diverse cohorts of general heart failure patients. Eur J Heart Fail. 2018;20(1):67–75.
18. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718.
19. Zhang P, Li S, Chen Z, Lu Y, Zhang H. LncRNA SNHG8 promotes proliferation and invasion of gastric cancer cells by targeting the miR-491/PDGFRA axis. Hum Cell. 2020;33(1):123–130.
20. Meng Q, Wang X, Xue T, Zhao Q, Wang W, Zhao K. Long non-coding RNA MIR99AHG promotes gastric cancer progression by inducing EMT and inhibiting apoptosis via miR577/FOXP1 axis. Cancer Cell Int. 2020;20:414.
21. Zheng S, Jiang F, Ge D, et al. LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed Pharmacother/Biomedecine Pharmacotherapie. 2019;112:108695.
22. Bi S, Liu Z, Wu Z, et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 2020;11(7):483–504.
23. Tang X, Shi L, Xie N, et al. SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat Commun. 2017;8(1):318.
24. Lu YF, Xu XP, Lu XP, et al. SIRT7 activates p53 by enhancing PCAF-mediated MDM2 degradation to arrest the cell cycle. Oncogene. 2020;39(24):4650–4665.
25. Wei W, Jing ZX, Ke Z, Yi P. Sirtuin 7 plays an oncogenic role in human osteosarcoma via downregulating CDC4 expression. Am J Cancer Res. 2017;7(9):1788–1803.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.