Back to Journals » OncoTargets and Therapy » Volume 13
LINC02738 Participates in the Development of Kidney Cancer Through the miR-20b/Sox4 Axis
Authors Han C, Xu B, Zhou L, Li L, Lu C, Yu GP, Liu YS
Received 18 June 2020
Accepted for publication 21 August 2020
Published 9 October 2020 Volume 2020:13 Pages 10185—10196
DOI https://doi.org/10.2147/OTT.S262046
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Chao Han,* Bin Xu,* Lin Zhou, Long Li, Chao Lu, Guo-Peng Yu, Yu-Shan Liu
Department of Urology, Shanghai Ninth People’s Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200011, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yu-Shan Liu
Department of Urology, Shanghai Ninth People’s Hospital, Shanghai Jiaotong University School of Medicine, No. 639 Zhizaoju Road, Shanghai 200011, People’s Republic of China
Tel +86-13701909308
Email [email protected]
Background: Long non-coding RNAs (lncRNAs) can affect tumorigenesis. Data from The Cancer Genome Atlas (TCAG) suggest that LINC02783 is highly expressed in renal cell carcinoma (RCC) and is expected to be a potential biological target. We conducted this study to verify this.
Patients and Methods: We conducted this study to verify the opinion that “LINC02783 is highly expressed in renal cell carcinoma (RCC) and is expected to be a potential biological target”. We employed quantitative real-time polymerase chain reaction (qRT-PCR) to test LINC02783 expression in RCC tissues, CKK-8 assay and transwell assay to assess the viability and invasion of RCC cells, Western blot to quantify Sox-4 expression, dual-luciferase reporter (DLR) assay and RNA immunoprecipitation (RIP) assay to analyze the interaction between LINC02783 and miR-20b, in vivo experiments to test tumor formation.
Results: We detected high LINC02783 expression in RCC patients. Patients with higher LINC02783 levels had a markedly poorer prognosis. In vitro and in vivo, the down-regulation of LINC02783 suppressed the viability and invasion of RCC cells. The DLR assay results revealed that LINC02783 enhanced Sox-4 expression by regulating miR-20b. LINC02783 can act as a sponge for miR-20b to inhibit Sox-4 expression.
Conclusion: LINC02783 is highly expressed in RCC patients and indicates a poor prognosis. LINC02783 can affect the occurrence and progression of RCC through the miR-20b/Sox-4 axis, making it a promising target for the treatment of RCC.
Keywords: LINC02783, miR-20b, Sox-4, renal cell carcinoma, The Cancer Genome Atlas, prognosis
Core Tip
In this study, LINC02783 was highly expressed in patients with RCC and caused a poor prognosis. LINC02783 can mediate the miR-20b/Sox-4 axis and regulate RCC cell growth, expected to become a potential target for RCC treatment.
Introduction
Renal cell carcinoma (RCC), occurring in the urinary system, is the second leading cause of deaths from malignant tumors in the urinary system.1 According to a global epidemiological survey,2 the number of kidney cancer patients in 2018 exceeded 400,000 and the number of deaths reached 170,000, accompanied by a yearly increase in the morbidity. RCC is divided into kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP) and kidney renal clear cell carcinoma (KIRC) according to the pathological type. KIRC is the most frequent RCC subtype (comprising 80–90% of RCC), characterized by high metastasis and recurrence rates.3–5 Advancements in medical levels have improved the prognosis of RCC patients. Recurrence and metastasis of RCC occur in over 30% of patients, leading to a median survival of about 13 months.6,7 Our exploration of the underlying mechanism of RCC will enlighten new strategies for RCC treatment.
A growing number of studies have realized the importance of non-coding RNAs (ncRNAs) in human tumors.8,9 Long non-coding RNA (lncRNA)s are transcribed RNAs with over 200 nucleotides but with no potential for protein-coding.10,11 By competing with miRs for microRNA response elements (MREs), lncRNAs inhibit the synthesis of miRs and thereby affect the expression of their downstream target genes.12,13 LncRNAs can regulate various cellular processes.14 The study by Feng et al15 suggests that the up-regulation of lncRNA MEG3 results in inhibited migration and invasion of bladder cancer cells and increased cisplatin-sensitivity. A former study indicates that16 lncRNA-SNHG16 stimulates tumor proliferation by epigenetically inhibiting p21 expression in bladder cancer and can predict the poor prognosis of patients. Many lncRNAs can regulate tumor progression and metastasis, but the function of lncRNAs in RCC remains unknown.17 No studies have been done to investigate LINC02783, a lncRNA located on human chromosome 11p15.3. We conducted a screening analysis of data from The Cancer Genome Atlas (TCGA) and noted high LINC02783 expression levels in KIRC, which suggests the potential of LINC02783 to become an indicator for the prognosis or diagnosis of KIRC.
Here we explored the clinical value and the underlying mechanism of LINC02783 in RCC, hoping to identify potential therapeutic targets for RCC treatment.
Materials and Methods
TCGA Data Download and Analysis
KIRC transcript data were downloaded from https://portal.gdc.cancer.gov/and then synthesized into a matrix file using the putFilesToOneDir.pl., followed by data analysis on Edger software package (logFC = 4, P = 0.05). Clinical data of KIRC patients were downloaded from http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/and synthesized using the Survival.pl. Then we subjected data to the Cox regression analysis and drew a survival curve.
Sample Collection
We enrolled 50 patients with RCC admitted to Shanghai Ninth People’s Hospital, Shanghai Jiaotong University School of Medicine from March 2012 to March 2014. We sampled cancerous tissues and adjacent tissues from patients and stored them in liquid-nitrogen at −80°C, followed by RNA detection. Patients were diagnosed with KIRC by the pathological test, with no history of anti-tumor treatments before, in line with the Fuhrman tumor grade criteria. This study was carried out with informed consent of all patients and ethical approval of the ethics committee of Shanghai Ninth People’s Hospital, Shanghai Jiaotong University School of Medicine and was in accordance with Helsinki Declaration (1536–08-65). Animal experiments were guided by the “Laboratory animal—Guideline for ethical review of animal welfare”.
Cell Culture
RCC cell lines (ACHN, 786-O, OSRC-2, and Caki-2) and immortalized normal human proximal tubular epithelial cell line (HK-2) were purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). RCC cells were cultured in a RPMI 1640 medium containing 10% fetal bovine serum (FBS) and HK-2 cells in keratinocyte-SFM (Gibco/Invitrogen, Vienna, Austria), together with 100 U/mL penicillin and 100 μg/mL streptomycin in a sterilized environment, with 5% CO2 at 37°C.
Cell Transfection
ACHN and 786-O cells at logarithmic growth phase were harvested and plated in a 6-well plate (1×106 cells/well). Transfection was done when cell confluency reached 90%. The pcDNA3.1 vector was used to construct the LINC02783 inhibitor (si-LINC02783), Sox-4 inhibitor (si-Sox-4), and Sox-4 overexpression vector (pcDNA-Sox-4). The design and synthesis of sequences of miR-20b-mimics and miR-20b-inhibit were completed by Sangon Biotech (Shanghai) Co., Ltd. The miR-NC worked as the control of miR, and si-NC as the control of other genes. Then we separately transfected cells with expression vectors, miR-20b-mimics, and miR-20b-inhibit according to the instructions of the Lipofectamine 2000 kit (Invitrogen™, USA).
Cell Viability Assay
CKK-8 assay (TransGen, Beijing, China) was used to test cell viability. Transfected cells were plated in a 96-well plate (5×103 cells/well) and cultured for 24 hours. Then the plate was incubated for 24, 48, and 72 hours. After incubation, 10 µL of CCK-8 solution was added to each well and the plate was incubated at 37°C for 2 hours. An automatic microplate reader (Bio-Rad, Hercules, CA, USA) was used to measure the absorbance at 450 nm.
Detection of Cell Invasion
Cell invasion was measured by the transwell assay. Cells transfected with plasmids were plated in a 6-well plate and incubated for 72 hours. Cells were resuspended in the serum-free medium to obtain the cell suspension (1×105 cells/mL) and inoculated into the apical chamber pre-coated with Matrigel (1:20, BD Corning). Then, 750μL of 10% FBS was added to the basolateral chamber and the whole transwell system was inoculated at 37°C for 12 hours, with 5% CO2. Cells were infiltrated with methanol for 20 minutes and stained with 0.1% crystal violet in a dark room. Finally, cells invading through the membrane were counted under a microscope.
Apoptosis Detection
Annexin V Apoptosis Detection Kit was used to test cell apoptosis (Becton Dickinson, NJ, USA). Transfected cells were stained with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) for 25 minutes. Then cell apoptosis was assessed by flow cytometry (BD CantoII).
Western Blot Analysis
Proteins were lysed by the RIPA buffer (Cell Signal Technology, Inc., MA, USA) and quantified by the BCA kit (Beyotime Biotechnology, Shanghai, China). After electrophoresis, proteins were transferred to a PVDF membrane (Millipore, Billerica, MA). Blocked with skim milk, the membrane was incubated with the primary antibody at 4°C for a whole night and then with the secondary antibody for 1 hour at room temperature. Color development was performed on the ECL chemiluminescence detection system (Thermo Fisher Scientific, Rochester, NY).
RNA Extraction and Real-Time qPCR
The total RNA was separated with a TRIzol kit (Invitrogen, USA) and then subjected to ultraviolet spectrophotometry analysis and agarose gel electrophoresis to test its purity, concentration, and integrity. Reverse transcription was conducted using theTaqManTM Reverse Transcription Kit (Invitrogen, USA) according to the kit manual to obtain the cDNA for subsequent experiments. PCR amplification was finished with PrimeScript RT Master Mix kit (Takara Bio, Japan). The amplification system was 20 μL in total, including 10 μL of SYBR qPCR Mix, 0.8 μL of upstream primer, 0.8 μL of downstream primer, 2 μL of cDNA product, 0.4 μL of 50×ROX reference dye, and RNase-free water to adjust the volume. PCR conditions: pre-denaturation at 95°C for 60s and then 40 cycles of denaturation at 95°C for 30s, annealing and extension at 60°C for 40s. We designed 3 parallel replicate wells and all specimens were tested three times. U6 and GADPH were used as internal references, and the data were analyzed using 2−ΔΔct.18 The PCR reaction was performed using the 7500 PCR instrument from ABI. Primer sequences are shown in Table 1.
 |
Table 1 Primer Sequences |
Dual-Luciferase Reporter (DLR) Assay
DNA oligonucleotides and pMiR-Reporter Vector were chosen for constructing LINC02783 wild/mutant type (LINC02783-WT/MUT) and Sox-4 wild/mutant type (Sox-4-WT/MUT). They were transfected into HEK293 cells, together with miR-150-5-mimics and miR-NC. Cells were collected after 24 hours of incubation and the fluorescence activity was tested using a DLR kit (Promega).
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted using the EZ-Magna RIP kit (Millipore, USA). RCC cells were lysed using RIPA. The whole-cell protein extract was incubated together with RIP wash buffer containing mouse immunoglobulin G (IgG) control or magnetic beads bound to the anti-AgO2 antibody (Millipool). Proteins were digested with proteinase K to isolate the immunoprecipitated RNA. The purified RNA was subjected to qRT-PCR to identify the presence of binding targets.
Models of Xenograft Tumor
Six-week-old BALB/c (nu/nu) nude mice purchased from Bena Culture Collection (Beijing) were housed in the barrier facility of the animal center of our hospital. Totally 5.0×106 RCC cells (786-O) with stably expressed sh-LINC02783 and sh-NC were transplanted to the right ribs of nude mice and measured the tumor volume in nude mice weekly. Tumor volume = 0.5 × width2 × length. After 28 days, nude mice were sacrificed to collect tumor tissues for testing. The animal experiment was conducted with approval of the Animal Ethics Committee of Shanghai Ninth People’s Hospital, Shanghai Jiaotong University School of Medicine (1695–12-86).
Statistical Analysis
The data visualization and analysis were performed on the GraphPad 7 software package. The analysis of independent prognostic factors was conducted on the SPSS20.0 software. The distribution of the measurement data was analyzed by the K–S test. Data with normal distribution were expressed by the mean ± standard deviation (Mean ± SD) and its intergroup comparison was analyzed by the independent sample t-test. The count data were expressed by the percentage (%) and its intergroup comparison was analyzed by the chi-square test, denoted by X2. The comparison between multiple groups was performed by one-way ANOVA and denoted by F. The LSD t-test was used for post hoc pairwise comparison. The comparison between multiple time points was performed by repeated measure ANOVA and denoted by F. The Bonferroni test was used for post hoc test. Pearson correlation coefficient was used to analyze the correlation between genes. Kaplan-Meier (K-M) survival curve was plotted to display the overall survival which was analyzed by the Log rank test. The difference was statistically significant when P<0.05.
Results
TCGA Data Analysis
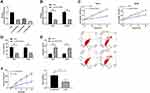
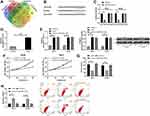
Data of 72 normal samples and 539 cancer samples from KIRC patients were downloaded from the TCGA database and subjected to a differential analysis. We found 112 differentially expressed lncRNAs, including 22 highly expressed ones and 90 lowly expressed ones (Figure 1A and B, Table 2). Cox regression analysis of clinical data found that age, AL357507.1, AC103925.1, AC093281.2, LUCAT1, and LINC02783 were independent prognostic factors affecting KIRC (Table 3).
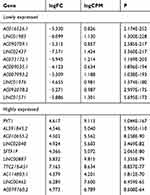 |
Table 2 Top 10 Lowly or Highly Expressed lncRNAs in KIRC |
 |
Table 3 Cox Regression Analysis |
Expression and Value of LINC02783 in KIRC Patients
Patients were divided into the high expression group and the low expression group by the median value of LINC02783 levels in the database. The results of overall survival favored patients with low LINC02783 expression over patients with high LINC02783 expression (Figure 2A). To further verify the expression of LINC02783 in KIRC, we tested the expression of LINC02783 in patient tissues. High LINC02783 expression was detected in cancer tissues and RCC cells (Figure 2B and C). Patients with high LINC02783 expression had larger tumor size (over 7 cm), advanced TNM stage (stage III + IV), and higher risks of tumor metastasis (Table 4). The 5-year survival greatly decreased in patients with high LINC00858 levels. Cox regression identified LINC00858 as an independent factor affecting the prognosis of patients (Table 5).
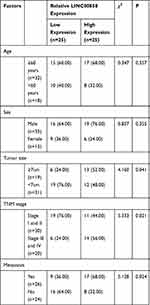 |
Table 4 Relationship Between LINC00858 and Low Pathological Data of Patients |
 |
Table 5 Cox Regression Analysis |
Knock-Down of LINC02783 Inhibited RCC Cell Viability and Invasion
Three LINC02783 interference sequences (si-LINC02783 # 1, # 2, # 3) were designed to further explore the role of LINC02783 in the biological function of RCC cells (Figure 3A). Si-LINC02783#3 (hereafter referred to as si-LINC02783) with the most differential expression was transfected into 786-O and ACHN cells, respectively (Figure 3B). The results of CCK-8 assay revealed that cells with si-LINC02783 had remarkably weaker viability compared with cells si-NC (Figure 3C). Transwell assay results found that the number of cells migrating through the membrane was lower in cells with si-LINC02783 than in cells with si- NC (Figure 3D). Flow cytometry results demonstrated that cells with si-LINC02783 had markedly higher apoptosis compared with cell with si-NC (Figure 3E). The tumor volume and weight were markedly lower in nude mice injected with sh-LINC02783 than in mice transfected with sh-NC (Figure 3F). We drew a conclusion from the above results that the knock-down of LINC02783 can inhibit the growth of RCC.
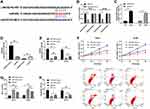
LINC02783 Acted as a miR-20b Sponge
MiRDB was used to predict the potential targeted miRNAs of LINC02783 to further determine the relevant mechanism of LINC02783. We discovered targeted binding sites between miR-20b and LINC02783 (Figure 4A). The results of the DLR assay demonstrated that the fluorescence activity of LINC02783-WT was inhibited by miR-20b-mimics, but LINC02783-MUT was not affected (Figure 4B). The results of RIP assay revealed that both LINC02783 and miR-20b were precipitated by Ago2 antibodies (Figure 4C), which suggests a targeted relationship between miR-20b and LINC02783 (Figure 4D and E). The experimental results revealed that the cells with up-regulated miR-20b had markedly lower cell viability and invasion, and markedly higher apoptosis compared with cells with miR-NC, but the changes were reversed after the knock-down of miR-20b (Figure 4F–H).
miR-20b Targeted Sox-4 to Inhibit RCC Cell Viability and Invasion
We predicted the miR-20b target genes on Targetscan, miRDB, StarBase, and miRTarBase and identified 21 potential target genes (Figure 5A). Sox-4 was selected for further verification. DLR assay showed that miR-20b-mimics inhibited the fluorescence activity of Sox-4-WT but did not affect Sox-4-MUT (Figure 5B and C). The inhibition of Sox4 expression by si-Sox4 was reversed by miR-20b-mimics. The experimental results revealed that cells with Sox-4 knockdown had markedly lower cell viability and invasion, and markedly higher apoptosis than cells with si-NC, but these changes were reversed after the transfection with miR-20b-mimics (Figure 5F–H).
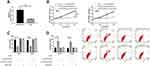
LINC02783 Inhibited RCC Cell Viability and Invasion via the miR-20b/Sox-4 Axis
To verify the results that LINC02783 could regulate the biological function of RCC cells via the miR-20b/Sox-4 axis, RCC cells were transfected with LINC02783, miR-20b, and Sox-4 and then their biological functions were assessed (Figure 6A). We found that the co-transfection of miR-20b-mimics, pcDNA-Sox-4 and si-LINC02783 reversed the suppression of RCC cell viability (Figure 6B) and invasion (Figure 6C) and the stimulation of apoptosis (Figure 6D) by si-LINC02783, which suggests that LINC02783 can regulate miR-20b/Sox-4 axis to regulate the biological functions of RCC.
miR-20b and Sox-4 Expression Levels in Patient Tissues and Their Correlation with LINC02783
At the end of the study, the relative expression of miR-20b and Sox-4 in patient tissues were tested. The test results revealed low miR-20b levels and increased Sox-4 levels in KIRC patient tissues (Figure 7A). Correlation analysis found that LINC02783 was in negative correlation with miR-20b and in positive correlation with Sox-4, while miR-20b and Sox-4 were negatively correlated (Figure 7B). The survival analysis showed that the 5-year survival was markedly lower in patients with low miR-20b expression and high Sox-4 expression (Figure 7C).
Discussion
Deaths caused by RCC are common worldwide, what’s worse, the morbidity of RCC is increasing year by year.19,20 LncRNAs, a type of non-coding RNAs, are closely related to RCC.21,22 In this study, we screened the potential differential lncRNAs of KIRC through the TCGA database and found that LINC02783 was highly expressed in KIRC and its high expression predicted poor survival. Such results are consistent with clinical findings. We also discovered that the knock-down of LINC02783 led to lower viability and invasion, as well as higher apoptosis in RCC cells, which was then confirmed by the tumor formation experiment in nude mice. These results indicate that LINC02783 may promote the occurrence and progression of RCC.
LncRNAs act as ceRNAs in biological processes. They can sponge miRNAs to regulate the expression of miRNA downstream target genes and affect tumor growth.23–25 The study by Zhao et al26 suggests that the lncRNA SNHG5/miR-32 axis can affect gastric cancer cell proliferation and migration through targeting KLF4. A previous study27 also indicates that lncRNA TUG1 affects the proliferation, migration, and the epithelial-mesenchymal transition (EMT) of papillary thyroid cancer cells by targeting miR-145. We conducted an online prediction of the potential target genes of LINC02783 on miRDB and found targeted binding sites between miR-20b and LINC02783. MiR-20b, located on the human chromosome Xq26.2, can suppress tumors and is lowly expressed in various tumors like lung cancer, glioma, and colorectal cancer.28–30 Here we detected low miR-20b expression levels in RCC patients and found that its low expression predicted a low survival rate of patients. Clinical detection revealed that miR-20b expression was negatively correlated with LINC02783 in patient tissues, which implies a possible correlation between LINC02783 and miR-20b. In order to verify the relationship between the two, we performed the DLR assay and RIP assay. It turned out that the fluorescence activity of LINC02783-WT was inhibited by miR-20b-mimics and both LINC02783 and miR-20b were precipitated by Ago2 protein, which indicates that LINC02783 can target miR-20b and act as a sponge for miR-20b. In this study, the up-regulation of miR-20b resulted in lower cell viability and invasion, and higher apoptosis, but those biological changes were reversed by the knock-down of miR-20b, suggesting that miR-20b is involved in RCC development and progression.
It is a classic mechanism that miRNAs regulate downstream target genes to affect biological processes.31 To figure out the specific mechanism of miR-20b, we conducted a joint prediction of the potential target genes of miR-20b on 4 different websites and identified 21 target genes. We selected Sox-4 for the following research according to literature and related information. Sox-4, as a member of the Sox family, can regulate embryonic development and cell growth.32–34 Sox-4 has a high expression level in RCC and boosts cell migration and invasion of RCC by inducing EMT.35 MiR-138 and miR-338-3p can affect the development of RCC by regulating Sox-4.36,37 Here we detected high Sox4 expression in the tissues of KIRC patients, which was positively correlated with LINC02783 expression and led to a poor prognosis. The DLR assay results suggest that miR-20b can also target Sox4. Cell experiments demonstrated that miR-20b-mimics reversed the inhibition of cell viability and invasion from the knock-down of Sox-4 and reversed the stimulation of cell apoptosis. Such results indicate that the miR-20b/Sox-4 axis can affect the progression of RCC. To verify that LINC02783 can inhibit the growth of RCC cells through the miR-20b/Sox-4 axis, we separately performed co-transfection and found that the inhibition of RCC cell growth by the knock-down of LINC02783 was reversed by miR-20b-inhibit and pcDNA-Sox-4.
Here we reveal that LINC02783 can inhibit the growth of RCC cells by regulating the miR-20b/Sox-4 axis, but this study is subject to certain limitations. Firstly, the small sample size may cause biases in the K-M survival analysis. Secondly, we did not explore the possibility of LINC02783 to work as a diagnostic indicator for RCC. Finally, here we only studied the effects of LINC02783 on RCC cells by regulating the miR-20b/Sox-4 axis, failing to explore other potential mechanisms of LINC02783 to affect the development of RCC. We will include more samples and perfect our study in the future to better support our study results.
Conclusion
This study found that LINC02783 was highly expressed in RCC patients and predicted a poor prognosis. LINC02783 can affect the occurrence and progression of RCC through the miR-20b/Sox-4 axis, making it a promising target for the treatment of RCC.
Supportive Foundations
This study didn’t receive any financial support.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356–387. doi:10.1016/j.ejca.2018.07.005
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Mouallem NE, Smith SC, Paul AK. Sarcomatoid renal cell carcinoma: biology and treatment advances. Urol Oncol. 2018;36(6):265–271. doi:10.1016/j.urolonc.2017.12.012
4. Vermassen T, De Meulenaere A, Van de Walle M, Rottey S. Therapeutic approaches in clear cell and non-clear cell renal cell carcinoma. Acta Clin Belg. 2017;72(1):12–18. doi:10.1080/17843286.2016.1193269
5. Akhtar M, Al-Bozom IA, Al Hussain T. Papillary Renal Cell Carcinoma (PRCC): an update. Adv Anat Pathol. 2019;26(2):124–132. doi:10.1097/PAP.0000000000000220
6. Godley PA, Ataga KI. Renal cell carcinoma. Curr Opin Oncol. 2000;12(3):260–264. doi:10.1097/00001622-200005000-00013
7. Ran L, Liang J, Deng X, Wu J. miRNAs in prediction of prognosis in clear cell renal cell carcinoma. Biomed Res Int. 2017;2017:4832931. doi:10.1155/2017/4832931
8. Li JK, Chen C, Liu JY, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16(1):111. doi:10.1186/s12943-017-0681-0
9. Zhai W, Sun Y, Guo C, et al. LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ. 2017;24(9):1502–1517. doi:10.1038/cdd.2017.74
10. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
11. Cai Z, Xu K, Li Y, Lv Y, Bao J, Qiao L. Long noncoding RNA in liver cancer stem cells. Discov Med. 2017;24(131):87–93.
12. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi:10.1158/2159-8290.CD-13-0202
13. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi:10.1016/j.canlet.2013.06.013
14. Yang Q, Wan Q, Zhang L, et al. Analysis of LncRNA expression in cell differentiation. RNA Biol. 2018;15(3):413–422. doi:10.1080/15476286.2018.1441665
15. Feng SQ, Zhang XY, Fan HT, Sun QJ, Zhang M. Up-regulation of LncRNA MEG3 inhibits cell migration and invasion and enhances cisplatin chemosensitivity in bladder cancer cells. Neoplasma. 2018;65(6):925–932. doi:10.4149/neo_2018_180125N55
16. Cao X, Xu J, Yue D. LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther. 2018;25(1–2):10–17. doi:10.1038/s41417-017-0006-x
17. St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239–251. doi:10.1016/j.tig.2015.03.007
18. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
19. Escudier B, Porta C, Schmidinger M, et al. Renal cell carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(suppl5):v58–v68. doi:10.1093/annonc/mdw328
20. Doehn C, Grunwald V, Steiner T, Follmann M, Rexer H, Krege S. The diagnosis, treatment, and follow-up of renal cell carcinoma. Dtsch Arztebl Int. 2016;113(35–36):590–596. doi:10.3238/arztebl.2016.0590
21. Hong Q, Li O, Zheng W, et al. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017;8(5):e2772. doi:10.1038/cddis.2017.181
22. Ye XT, Huang H, Huang WP, Hu WL. LncRNA THOR promotes human renal cell carcinoma cell growth. Biochem Biophys Res Commun. 2018;501(3):661–667. doi:10.1016/j.bbrc.2018.05.040
23. Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148–64167. doi:10.18632/oncotarget.11637
24. Zhang Z, Qian W, Wang S, et al. Analysis of lncRNA-associated ceRNA network reveals potential lncRNA biomarkers in human colon adenocarcinoma. Cell Physiol Biochem. 2018;49(5):1778–1791. doi:10.1159/000493623
25. Yin H, Wang X, Zhang X, et al. Integrated analysis of long noncoding RNA associated-competing endogenous RNA as prognostic biomarkers in clear cell renal carcinoma. Cancer Sci. 2018;109(10):3336–3349. doi:10.1111/cas.13778
26. Zhao L, Han T, Li Y, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31(3):893–903. doi:10.1096/fj.201600994R
27. Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin (Shanghai). 2017;49(7):588–597. doi:10.1093/abbs/gmx047
28. Leidinger P, Brefort T, Backes C, et al. High-throughput qRT-PCR validation of blood microRNAs in non-small cell lung cancer. Oncotarget. 2016;7(4):4611–4623. doi:10.18632/oncotarget.6566
29. Goze C, Reynes C, Forestier L, Sabatier R, Duffau H. Pilot study of whole blood MicroRNAs as potential tools for diffuse low-grade gliomas detection. Cell Mol Neurobiol. 2018;38(3):715–725. doi:10.1007/s10571-017-0536-7
30. Ulivi P, Canale M, Passardi A, et al. Circulating plasma levels of miR-20b, miR-29b and miR-155 as predictors of bevacizumab efficacy in patients with metastatic colorectal cancer. Int J Mol Sci. 2018;19(1):. doi:10.3390/ijms19010307
31. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015. 4. doi:10.7554/eLife.05005
32. Li J, Shen J, Wang K, Hornicek F, Duan Z. The roles of sox family genes in sarcoma. Curr Drug Targets. 2016;17(15):1761–1772. doi:10.2174/1389450117666160502145311
33. Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(1):15–30. doi:10.1016/j.stem.2012.12.007
34. Grimm D, Bauer J, Wise P, et al. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2019. doi:10.1016/j.semcancer.2019.03.004
35. Ruan H, Yang H, Wei H, et al. Overexpression of SOX4 promotes cell migration and invasion of renal cell carcinoma by inducing epithelial-mesenchymal transition. Int J Oncol. 2017;51(1):336–346. doi:10.3892/ijo.2017.4010
36. Tong Z, Meng X, Wang J, Wang L. MicroRNA-338-3p targets SOX4 and inhibits cell proliferation and invasion of renal cell carcinoma. Exp Ther Med. 2017;14(5):5200–5206. doi:10.3892/etm.2017.5169
37. Liu F, Wu L, Wang A, et al. MicroRNA-138 attenuates epithelial-to-mesenchymal transition by targeting SOX4 in clear cell renal cell carcinoma. Am J Transl Res. 2017;9(8):3611–3622.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.