Back to Journals » OncoTargets and Therapy » Volume 13
LINC01572 Regulates Cisplatin Resistance in Gastric Cancer Cells by Mediating miR-497-5p
Authors Song Z, Jia N, Li W, Zhang XY
Received 16 June 2020
Accepted for publication 18 September 2020
Published 27 October 2020 Volume 2020:13 Pages 10877—10887
DOI https://doi.org/10.2147/OTT.S267915
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Nicola Silvestris
Zhe Song,1 Nan Jia,1 Wei Li,1 Xiao-Yu Zhang2
1Second Department of General Surgery, Cangzhou Central Hospital, Cangzhou, Hebei Province, Mainland China; 2Department of Thyroid and Breast III, Cangzhou Central Hospital, Cangzhou, Hebei Province, Mainland China
Correspondence: Xiao-Yu Zhang
Department of Thyroid and Breast III, Cangzhou Central Hospital, Cangzhou, Hebei Province, People’s Republic of China
Email [email protected]
Background: Chemotherapy resistance has long been recognized as a major obstacle to cancer treatment. Therefore, elucidating the underlying mechanisms of chemotherapy resistance is conducive to developing new strategies to improve patients’ response to chemotherapy drugs.
Materials and Methods: Real-time quantitative PCR (QPCR) was applied to measure the expression levels of lncRNAs. LINC01572 was down-regulated or up-regulated in GC cells transfected with either LINC01572 shRNA or overexpression vectors. In vitro and in vivo experiments were conducted to investigate the role of LINC01572 in autophagy-related chemotherapy resistance.
Results: Compared with the parental cells, drug-resistant GC cells had a higher level of LINC01572. Silencing of LINC01572 inhibited chemotherapy-induced autophagy, while its knockout sensitized GC cells against chemotherapy drugs. As a competitive endogenous RNA of miR-497-5p, LINC01572 weakened the inhibitory effect of miR-497-5p on ATG14, leading to chemically induced autophagy and chemotherapy resistance in GC cells.
Conclusion: A new mechanism of GC autophagy-related chemotherapy resistance regulated by lncRNA was explored in this study, providing a new perspective for understanding chemotherapy resistance.
Keywords: LINC01572, miR-497-5p, gastric cancer, drug resistance
Introduction
As the most frequent and prevalent malignant gastrointestinal tumor globally, gastric cancer (GC) has been reported to be the third leading cause of cancer-related deaths.1,2 Although the continuous improvement of medical standards has brought about a great leap in the treatment and diagnosis of GC in various aspects, most advanced GC patients still confronted with the fact that the disease cannot be cured, and the prognosis is extremely unsatisfactory.3 Studies have found that4 perioperative adjuvant chemotherapy can effectively reduce distal metastasis and improve the survival rate of GC patients. Clinically, cisplatin (DDP) is the mainstream chemotherapy drug for GC at present. However, in the process of chemotherapy, drug resistance is inevitable, which is a key obstacle to ameliorate the prognosis of GC patients.5,6 Hence, we hope to further explore new potential therapeutic targets to fulfill our ends of improving the survival and prognosis of GC patients by understanding the underlying mechanism.
Belonging to the family of endogenous non-coding RNA,7 long non-coding RNA (lncRNA) is reported be more than 200nt in length with the capacity to encode protein, which is considered as an mRNA-like transcript.8 In recent years, studies have exhibited that9–11 lncRNAs participate in multiple biological functions during chromatin imprinting, chromatin modification, chromatin, and maintenance. What’s more, it is reported that12–15 lncRNAs are involved in the occurrence, development, invasion, and metastasis of tumors, and is closely related to drug resistance of tumors. For example, Zhang et al16 revealed that PVT1 overexpression enhanced multi-drug resistance in GC cells, and other evidence has17 exhibited that overexpression of GHET1 increased multi-drug resistance of GC cells. All these indicate that lncRNA is closely related to GC resistance. Of these, LINC01572, a member of the lncRNA family, has been shown to be differentially expressed in lung cancer expression profile,18 but its role in GC has not been clarified. In order to determine the expression of LINC01572 in GC, we search TCGA database to find that it was highly expressed in GC patients, which suggested its role as a potential diagnosis of GC. However, whether it can be a target of drug resistance in GC needs further exploration.
Therefore, this study was set out to explore the clinical value of LINC01572 in GC and the mechanism related to drug resistance, in an attempt to provide a potential target for clinical treatment of this disease.
Materials and Methods
Tissue Sample Collection
Thirty patients with GC, all resistant to DDP, who were treated in our hospital from May 2014 to March 2017 were enrolled (drug resistance group), and another 30 patients with GC who were sensitive to DDP treatment (non-drug resistance group) were collected. All patients underwent surgical treatment in our hospital. The tumor tissues of the patients were collected during the operation and stored at −80 °C for later use. Concurrently, 30 healthy controls were collected as the control group. Peripheral blood of the three groups was collected to detect the expression of LINC01572. This study was approved by the Medical Ethics Committee of Cangzhou Central Hospital, and written informed consent was obtained from each participant.
Cell Line Source
Normal gastric mucosal cell line GES-1 and parental GC cell lines BGC823 and SGC7901, as well as corresponding DDP-resistant strains (BGC823/DDP and SGC7901/DDP) were all purchased from the American Type Culture Collection (ATCC; Rockville, MD, USA). All the cells were cultured in PRMI-1640 medium (Gibco, USA) containing 10% bovine fetal serum (Gibco, USA) 37 °C with 5% CO2.
Cell Transfection
Cell transfection was conducted with the Lipofectamine 3000 (Invitrogen) kit. pcDNA3.1 was adopted to construct the LINC01572 overexpression and inhibition vectors (pcDNA3.1-LINC01572, si-LINC01572), and the empty vector pcDNA3.1 (Vector) was used as the control. Besides, miR-497-5p overexpression (miR-497-5p-mimics), miR-497-5p inhibitor (miR-497-5p-inhibit) and negative control (miR-NC) were established.
PCR Detection
Total RNA was extracted from patients’ serum, nude mice tissues and cells by TRIzol kit (Invitrogen, USA), and the determination of its purity, concentration, and integrity was conducted by UV spectrophotometer and agarose-gel electrophoresis. Subsequently, TaqMan ™ Reverse Transcription Kit (Invitrogen, USA) was applied for reverse transcription strictly following the kit instructions. The obtained cDNA was reserved for subsequent research. Then came the PCR amplification, which was performed with the help of PrimeScript RT Master Mix kit (Takara Bio, Japan). Amplification system: SYBR qPCR Mix: 10 μL, upstream primer: 0.8 μL, downstream primers: 0.8 μL, cDNA product: 2 μL, 50 × ROX reference dye: 0.4 μL, and RNase-free water in a final volume of 20 μL. PCR reaction conditions (40 cycles): pre-denaturation: 95 °C, 60s, denaturation: 95 °C, 30s, annealing and extension: 60 °C, 40s. Three parallel replicate wells were designed in the experiment, and all specimens were tested 3 times. U6 and GADPH were used as the internal parameters, and 2− ΔΔ ct was utilized for data analysis.19 The PCR instrument used in this study was 7900 PCR from ABI.
Western Blot Detection
The collected cells were lysed with RIPA lysate (Thermo Scientific™, USA), and centrifuged for 10min at 16,000*g to prepare the homogenate. Then, the proteins were quantified using the BCA kit (Thermo Scientific™, USA). Subsequently, the proteins were sealed with 5% skim milk, and added with primary antibodies ATG14, LC3I/II, and β-actin (R&D, USA) at a ratio of 1:1000, for a 90-minute incubation. Thereafter, horseradish peroxidase-coupled goat anti-mouse/rabbit IgG (R&D, USA) was added as the secondary antibody (1:1000). Finally, the Western blot was detected using enhanced chemiluminescence reagent (ECL; Thermo Fisher Scientific, USA), and Amersham Prime ECL Plus detection system was used for detection.
DDP Resistance Detection
Cell resistance detection was performed by the MTT method. First, the viability of CG cells exposed to DDP at different concentrations (0.5, 1, 2, 4, 8, 16, 32 μ/mL) was measured. Then, the IC50 value (half maximum inhibitory concentration) was used to determine the DDP sensitivity, and the inhibition rate of cells treated with varying concentrations of DDP was calculated to draw the inhibition curve. Note: The concentration of DDP in the curve corresponding to 50% inhibition of cell viability was the IC50 value.
Apoptosis Detection
Cell apoptosis and cell cycle were determined by Flow cytometry, specifically as follows: after adjusting to a cell suspension of 1×106, the cells were seeded in cell culture flasks for overnight growth after 48hours of transfection. Then, the cells were collected and rinsed with PBS, and the apoptosis rate was measured using Annexin V-FITC apoptosis detection kit (Invitrogen ™, USA) following the kit instructions.
Cell Invasion Experiment
Transwell chamber was used to detect cell invasion, and the specific steps were as follows: BGC823/DDP and SGC7901/DDP were adjusted to 1×104 and seeded into Transwell upper chamber where 50 ul Matrigel was added, and the lower chamber was added with 600 mL FBS (20%). After 48 hours of incubation, the invading cells were immobilized with 4% paraformaldehyde for 30min before washing with PBS, followed by the staining with 0.1% crystal violet for 15min. At last, the number of cell invasion was counted under a microscope.
Dual-Luciferase Reporter Assay
The potential targets of LINC01572 and miR-497-5p were predicted by StarBase v3.0 and TargetScan, respectively, and verified by luciferase reporter gene assay. The synthesized 3ʹ-UTR sequence or mutant UTR sequence of LINC01572 and ATG14 were then inserted into the pGL3 control vector (Promega, Madison, Wisconsin, USA). Thereafter, HEK-293T and miR-497-5p were transfected with wild-type or mutant LINC01572-3ʹ-UTR and ATG14-3ʹ-UTR, respectively, followed by the transduction with miR-497-5p mimics. Forty-eight hours later, the luciferase activity of the collected cells was measured using a dual-luciferase reporter kit (Berthold, Bad Wildbad, Germany).
RIP
RNA immunoprecipitation (RIP) was used to identify whether LINC01572 and miR-497-5p could interact with or bind to Ago2, a possible binding protein in BGC823 cells. EZMagna RIP kit (Millipore, Billerica, MA, USA) was applied according to the manufacturer’s instructions. The EC109 cells were lysed and incubated with protein A magnetic beads, which were conjugated with antibodies at 4 °C. Six hours later, the beads were washed with washing buffer and then incubated with 0.1% SDS and 0.5 mg/mL proteinase K at 55 °C for 30 minutes to remove the protein. Finally, qRT-PCR analysis was performed on the immunoprecipitated RNA to demonstrate the presence of LINC01572 and miR-497-5p using specific primers.
Tumorigenesis Experiment in Nude Mice
Twelve 4-week-old BALB/c nude mice, with a mass of 8–10g, were purchased from Beijing Vital RiverLaboratories Co., Ltd and subdivided into four groups, with 3 mice in each group. Together with and stably transfected BGC823/DDP cells (1.0×107), Vector, miR-497-5p-mimics+Vector, pcDNA3.1-LINC01572+Vector, pcDNA3.1-LINC01572+miR-497-5p-mimics, were respectively injected into the right side of the posterior teeth of nude mice. The experiment lasted for 4 weeks. The nude mice were injected intraperitoneally with DDP PBS (10 mg/kg) since the 2nd week, and the tumor volume (formula: 0.5 × length × width2) of nude mice was measured once a week. Four weeks later, all the nude mice were killed to weigh the tumor mass. The animal experimental procedures were approved by the animal experimental ethics committee of Cangzhou Central Hospital Cangzhou Central Hospital, and the experiments were carried out following the Guide for the Care and Use of Experimental Animals.
Statistical Analysis
In this study, the collected data was statistically analyzed using SPSS20.0, and the picture rendering was performed by GraphPad 7. The K–S test was used to analyze the distribution of the dose data, among which the normal distribution data was represented by mean ± standard deviation (Mean ± SD). The independent sample t-test was adopted for inter-group comparison, and one-way analysis of variance (ANOVA) was employed for comparison among groups, expressed as F. The post hoc pairwise comparison was done by LSD-t test, and the multi-time expressions were analyzed by repeated measures ANOVA, denoted by F. Bonferroni was responsible for post hoc test, and the Pearson test was used to analyze the correlation of LINC01572 with miR-497-5p and ATG14 in patient tissues. P<0.05 indicated a statistically significant difference.
Results
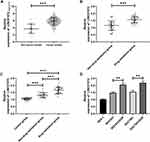
Overexpression of LINC01572 in DDP-Resistant GC Patients
We found through TCGA database that LINC01572 was highly expressed in cancer samples (Figure 1A). Then, we test its expression in DDP-resistant and non-drug-resistant patients through clinical experiments. It was observed that the expression of LINC01572 in non-DDP-resistant patients was markedly increased, as compared with DDP-resistant patients, while its expression in serum of DDP-resistant and non-resistant patients was remarkably elevated as compared to controls (Figure 1B and C). Besides, the expression of LINC01572 in drug-resistant cell lines was higher than that of parental cells and normal cells (Figure 1D). Overall, we confirmed that LINC01572 was highly expressed in GC, and was more significantly expressed in DDP-resistant GC patients.
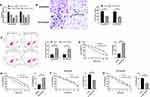
Knockout of LINC01572 Promoted Apoptosis and Invasion of DDP-Resistant Cell Lines and Reduced Drug Resistance
Through the above studies, we have confirmed that the expression of LINC01572 in drug-resistant GC cell lines was higher than that in parental cell lines. In order to observe the effect of LINC01572 on drug-resistant cell lines, we constructed three inhibitory sequences. It was found that si-LINC01572 # 2 had the most obvious inhibitory effect on DDP-resistant cell lines (Figure 2A). Subsequently, we detected the invasion and apoptosis of DDP cell lines transfected with si-LINC01572 #2. The results demonstrated that the number of BGC823/DDP and SGC7901/DDP invasion cells declined remarkably and the apoptosis rate increased after LINC01572 knockout (Figure 2B and C), suggesting that the decrease of LINC01572 could inhibit DDP-resistant cells’invasion. What’s more, the IC 50 of DDP-resistant cell lines decreased significantly after LINC01572 knockdown, which increased the sensitivity of DDP-resistant cell lines (Figure 2D–G).
LINC01572 Functioned as a Sponge for miR-497-5p
To determine the correlation between of LINC01572 disorders and DDP resistance, we further predicted the potential target of LINC01572 through the online software starbase3.0 (http://starbase.sysu.edu.cn/). It was found that miR-497-5p and LINC01572 had targeted binding sites (Figure 3A). Then, dual-luciferase activity was detected to confirm their relationship. It showed that the luciferase activity of LINC01572-WT was profoundly inhibited after co-transfection with miR-497-5p-mimics, while the luciferase activity of LINC01572 was not significantly changed after co-transfection with LINC01572-MUT, indicating that LINC01572 could bind to miR-497-5p in a targeted manner (Figure 3B). RIP experiment was further carried out to validate the correlation between the two. The results exhibited that the expression levels of LINC01572 and miR-497-5p precipitated by Ago2 antibodies were significantly higher than those precipitated by IgG (Figure 3C). Subsequently, we detected miR-497-5p expression in LINC01572 transfected cells. The results demonstrated that the miR-497-5p level in BGC823/DDP and SGC7901/DDP cells was markedly inhibited after up-regulation of LINC01572, while reverse results were obtained after LINC01572 knockout. Moreover (Figure 3D), miR-497-5p was found to be underexpressed in cancer tissues of DDP-resistant patients and was negatively correlated with LINC01572 (Figure 3E–G).
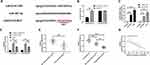
Up-Regulation of miR-497-5p Blocked ATG14, Inhibited Autophagy, and Promoted Apoptosis in DDP-Resistant Cells
To investigate the effect of miR-497-5p on cell resistance, we constructed miR-497-5p-mimics, miR-497-5p-inhibit, and the miR-NCs of BGC823/DDP and miR-NCSGC7901/DDP cells. It was found that miR-497-5p expression was notably increased after transfection with miR-497-5p-mimics (Figure 4C), the IC50 was significantly reduced, and the apoptosis rate increased as compared with miR-NC transected cells. The results of miR-497-5p-inhibit transfected cells were opposite to those of miR-497-5p-mimics transfected ones, that is, the expression of miR-497-5p declined markedly, the IC 50 increased, and the apoptosis rate decreased (Figure 4D–F). Then, to inquiry into the mechanism of miR-497-5p in drug resistance, we found through online prediction website Targetscan (http://www.targetscan.org/vert_72/) that ATG14 and miR-497-5p had targeted binding sites (Figure 4A). Later, dual-luciferase reporter analysis exhibited that the luciferase activity was markedly inhibited after co-transfection of ATG14-WT and miR-497-5p-mimics, while that of ATG14-MUT was not suppressed. Furthermore (Figure 4B), WB revealed that ATG14 was dramatically reduced after miR-497-5p-mimics transfection, and we also noticed the increase of LC3-I expression and decrease of LC3-II. Conversely, the transfection of miR-497-5p-inhibit resulted in the opposite results (Figure 4G). Moreover, the detection of clinical samples exhibited an increase in ATG14 expression in the tissues of drug-resistant patients. Correlation analysis confirmed that ATG14 was negatively related to miR-497-5p in patients’ tissues, and positively correlated with LINC01572 (Figure 5A–D).
 |
Figure 5 Expression and correlation analysis of Atg14 in GC. (A and B) ATG14 expression in patients with GC. (C and D) Correlation of ATG14 with miR-497-5p and LINC01572. ***Indicates P<0.001. |
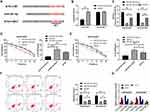
Down-Regulation of LINC01572 Inhibited Autophagy and Promoted Tumor Growth Through miR-497-5p ATG14 Axis
Vector, miR-497-5p-mimics+Vector, pcDNA3.1-LINC01572+Vector, pcDNA3.1-LINC01572+miR-497-5p-mimics nude mouse models were established to further observe whether LINC01572 affected GC growth through the miR-497-5p/ATG14 axis. After treating with DDP in the second week, the tumor volume and mass of nude mice injected with miR-497-5p-mimics+Vector were found to be significantly reduced (Figure 6A and (B); the detection of protein expression revealed that ATG14 and LC3-II were reduced, and LC3-I expression increased (Figure 6E). Also, we observed that the tumor volume and mass of nude mice after co-transfection of up-regulated LINC01572+miR-497-5p-mimics increased significantly compared with those merely transfected with miR-497-5p-mimics, and the protein expression reversed (Figure 6C and D). All these suggested that LINC01572 down-regulation inhibited autophagy and promoted the growth of GC cells by regulating miR-497-5p/ATG14 axis.
Discussion
Drug resistance in tumor therapy has always been a common clinical phenomenon, which seriously affects the treatment and prognosis of patients. This study revealed that LINC01572 expression was remarkably enhanced in patients with DDP-resistant GC. Further basic experiments revealed that LINC01572 can cause autophagy and make cells resistant to DDP by regulating the miR-497-5p/ATG14 axis, which was a promising clinical therapeutic target.
The occurrence of drug resistance in cancer patients is due to the long-term use of a sing drug, which makes tumor cells resistant to the drug.20 This is also the reason for the failure of tumor chemotherapy in clinical practice.21 Therefore, finding out the relevant mechanism of tumor drug resistance is the key to solve this problem. As a kind of long non-coding RNA, LncRNA has been reported to affect the occurrence of tumors, cardiovascular diseases, neurological diseases, and other diseases.22–24 Among them, it was found that25 lncRNA, as a competitive response element between sponge and miR, blocks its transcription and results in different expression. For example, Yan et al26 reported that by targeting miR-126 to activate the PI3K/AKT/MRP1 gene, lncRNA HOTAIR was able to promote DDP-resistance in GC. In addition, Wang et al27 revealed that lncRNA SNHG12 promoted multi-drug resistance of lung cancer by activating MAPK/slug pathway through sponging miR-181a. As a vital member of lncRNA family, LINC01572 is located on human 16q22.2 chromosome. Previous studies have found that18,28 LINC01572 is differentially expressed in polycystic ovary syndrome and lung cancer. The analysis of TCGA database in the present study exhibited that LINC01572 was highly expressed in GC, which suggested that LINC01572 might be implicated in the occurrence and development of GC. In light of this, we further carried out clinical and basic research.
Firstly, we found that healthy controls presented high LINC01572 expression compared with GC patients, and the LINC01572 expression in tissues of drug-resistant GC patients was markedly higher than that in non-resistant GC patients. All these findings were consistent with the database results, suggesting that LINC01572 participated in GC progression, and there was also an association between LINC01572 and GC resistance. Further, we conducted experiments in order to prove that LINC01572 affected the resistance of GC cells. It was found that after LINC01572 knockout, the invasion rate of cells was markedly reduced, the apoptosis rate was significantly elevated, and the IC50 of drug-resistant cells was notably decreased, indicating that LINC01572 knockout could improve the sensitivity of drug-resistant cell lines to DDP and had a sensitization effect. However, its related mechanism remains to be investigated. In this regard, we explored the relationship between the two and found the possible targeted binding sites between them through online bioinformatics analysis. MiR-497-5p is a tumor suppressor gene that presents low expression in many tumors such as breast cancer, lung cancer, and glioma.29–31 Feng et al32 also observed that miR-497-5p hindered the proliferation and growth of GC cells by targeting PDK3. However, whether miR-497-5p expresses differently in patients with GC resistance has not been verified. In our study, miR-497-5p was lowly expressed in GC patients, which accorded with the results of Liu et al.33 In addition, we confirmed that the expression level of miR-497-5p in drug-resistant patients was lower than that in non-drug-resistant patients. Subsequently, correlation analysis also demonstrated that miR-497-5p was negatively related to LINC01572 in patients’ tissues, indicating that miR-497-5p and LINC01572 might have a regulatory relationship. Then, dual-luciferase activity detection and RIP experiment were carried out to further validate that LINC01572 and miR-497-5p did have a targeted regulatory effect.
Autophagy refers to the manifestation of cell self-phagocytosis. Previously, it is reported that blocked autophagy may promote tumorigenesis.34 Other scholars have confirmed that35 autophagy can protect cells from stress responses caused by external environments such as starvation and chemoradiation. It is also well established that36 chemotherapy can induce autophagy in tumor cells, help tumor cells escape, avoid fatal injuries, and develop drug resistance in cells. ATG14 is an important autophagic factor that plays two roles in autophagy:37 one is to form a type III Ptdlns3k complex I as a key component, which is essential for the formation of autophagy; the other is to direct the complex I to the phage assembly site to play a role in autophagy. In the current study, the target binding sites between miR-497-5p and ATG14 was ascertained through online target gene prediction analysis, and was later confirmed by dual-luciferase activity experiment. Then, cell experiments were carried out to observe whether miR-497-5p regulated ATG14 to promote the occurrence of autophagy and induce cell resistance. The results showed that miR-497-5p up-regulation notably decreased the IC50 of cells and increased the apoptosis rate, enhanced the expression of ATG14 and LC3-I proteins, and reduced the expression of LC3-II proteins. On the contrary, the effects of inhibition of miR-497-5p were reversed. Subsequently, clinical research exhibited that ATG14 was highly expressed in patients with gastric cancer, and its expression was more significantly increased in patients with drug resistance. Through correlation analysis, we found that ATG14 was positively correlated with LINC01572, and negatively correlated with miR-497-5p, suggesting that LINC01572 and miR-497-5p/ATG14 might play an important regulatory role in GC.
At the end of the study, we injected co-transfected vectors into nude mice through in vivo experiments, and treated them with DDP at the second week of modeling, so as to verify the relationship between LINC01572 and miR-497-5p/ATG14. The results demonstrated that the tumor volume and mass of nude mice injected with miR-497-5p-mimics+Vector were significantly reduced, the expression levels of ATG14 and LC3-II were decreased, and the LC3-I was increased. While the tumor volume and mass of nude mice injected with LINC01572+ miR-497-5p-mimics were remarkably increased and the protein expression was reversed as compared with those transfected merely with miR-497-5p-mimics. Based on the above results, we can confirm that LINC01572 can induce autophagy in cells by regulating miR-497-5p/ATG14 axis, leading to cell resistance to DDP.
There are still some limitations in this study. Firstly, the relationship between LINC01572 and prognosis of gastric cancer patients remains to be studied. Secondly, although our study found that LINC01572 was highly expressed in GC patients, more clinical samples should be collected to verify whether it can be a potential diagnostic indicator. Therefore, we hope to collect more research samples in future studies and carry out follow-up to supplement our findings.
All in all, LINC01572 showed increased expression in DDP-resistant GC patients and can induce autophagy in cells by regulating the miR-497-5p/ATG14 axis, making cells resistant to DDP.
Data Sharing Statement
Derived data supporting the findings of this study are available from the corresponding author on request.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wang FH, Shen L, Li J, et al. The Chinese Society of Clinical Oncology (CSCO): clinical guidelines for the diagnosis and treatment of gastric cancer. Cancer Commun (Lond). 2019;39(1):10. doi:10.1186/s40880-019-0349-9
2. Banks M, Graham D, Jansen M, et al. British society of gastroenterology guidelines on the diagnosis and management of patients at risk of gastric adenocarcinoma. Gut. 2019;68(9):1545–1575. doi:10.1136/gutjnl-2018-318126
3. van den Ende T, Ter Veer E, Mali RMA, et al. Prognostic and predictive factors for the curative treatment of esophageal and gastric cancer in randomized controlled trials: a systematic review and meta-analysis. Cancers (Basel). 2019;11(4):530. doi:10.3390/cancers11040530
4. Cats A, Jansen EPM, van Grieken NCT, et al. Chemotherapy versus chemoradiotherapy after surgery and preoperative chemotherapy for resectable gastric cancer (CRITICS): an international, open-label, randomised Phase 3 trial. Lancet Oncol. 2018;19(5):616–628. doi:10.1016/S1470-2045(18)30132-3
5. Ye Y, Yang S, Han Y, et al. HOXD-AS1 confers cisplatin resistance in gastric cancer through epigenetically silencing PDCD4 via recruiting EZH2. Open Biol. 2019;9(9):190068. doi:10.1098/rsob.190068
6. Lu L, Wu M, Lu Y, et al. MicroRNA-424 regulates cisplatin resistance of gastric cancer by targeting SMURF1 based on GEO database and primary validation in human gastric cancer tissues. Onco Targets Ther. 2019;12:7623–7636. doi:10.2147/OTT.S208275
7. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47.
8. Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17(10):601. doi:10.1038/nrg.2016.85
9. Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12(1):1.
10. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
11. Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 2016;14(2):73–80. doi:10.1016/j.gpb.2016.03.005
12. Yoshimoto R, Mayeda A, Yoshida M, et al. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta Gene Regul Mech. 2016;1859(1):192–199. doi:10.1016/j.bbagrm.2015.09.012
13. Qi P, Zhou X, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15(1):39. doi:10.1186/s12943-016-0524-4
14. Raveh E, Matouk IJ, Gilon M, et al. The H19 Long non-coding RNA in cancer initiation, progression and metastasis–a proposed unifying theory. Mol Cancer. 2015;14(1):184. doi:10.1186/s12943-015-0458-2
15. Chandra Gupta S, NandanTripathi Y. Potential of long non‐coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–1967. doi:10.1002/ijc.30546
16. Zhang X, Bu P, Liu L, et al. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462(3):227–232. doi:10.1016/j.bbrc.2015.04.121
17. Zhang X, Bo P, Liu L, et al. Overexpression of long non-coding RNA GHET1 promotes the development of multidrug resistance in gastric cancer cells. Biomed Pharmacother. 2017;92:580–585. doi:10.1016/j.biopha.2017.04.111
18. Chen WJ, Tang RX, He RQ, et al. Clinical roles of the aberrantly expressed lncRNAs in Li X, Wang Q, Rui Y, et al. HOXC13‐AS promotes breast cancer cell growth through regulating miR‐497‐5p/PTEN axis. Journal of cellular physiology, 2019. Squamous cell carcinoma: a study based on RNA-sequencing and microarray data mining. Oncotarget. 2017;8(37):61282.
19. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
20. Chen C, Tang X, Liu Y, Zhu J, Liu J. Induction/reversal of drug resistance in gastric cancer by non-coding RNAs (Review). Int J Oncol. 2019;54(5):1511–1524. doi:10.3892/ijo.2019.4751
21. Satoh T, Lee KH, Rha SY, et al. Randomized Phase II trial of nimotuzumab plus irinotecan versus irinotecan alone as second-line therapy for patients with advanced gastric cancer. Gastric Cancer. 2015;18(4):824–832. doi:10.1007/s10120-014-0420-9
22. Chi Y, Wang D, Wang J, Yu W, Yang J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells. 2019;8(9):1015. doi:10.3390/cells8091015
23. Greco S, Gaetano C, Martelli F. Long noncoding competing endogenous RNA networks in age-associated cardiovascular diseases. Int J Mol Sci. 2019;20(12):3079. doi:10.3390/ijms20123079
24. Chen Y, Zhou J. LncRNAs: macromolecules with big roles in neurobiology and neurological diseases. Metab Brain Dis. 2017;32(2):281–291. doi:10.1007/s11011-017-9965-8
25. Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148. doi:10.18632/oncotarget.11637
26. Yan J, Dang Y, Liu S, et al. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biol. 2016;37(12):16345–16355. doi:10.1007/s13277-016-5448-5
27. Wang P, Chen D, Ma H, et al. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017;8(48):84086. doi:10.18632/oncotarget.20475
28. Zhao J, Xu J, Wang W, et al. Long non-coding RNA LINC-01572: 28 inhibits granulosa cell growth via a decrease in p27 (Kip1) degradation in patients with polycystic ovary syndrome. EBioMedicine. 2018;36:526–538. doi:10.1016/j.ebiom.2018.09.043
29. Li X, Wang Q, Rui Y, et al. HOXC13‐AS promotes breast cancer cell growth through regulating miR‐497‐5p/PTEN axis. J Cell Physiol. 2019;234:22343–22351.
30. Huang X, Wang L, Liu W, Li F. MicroRNA-497-5p inhibits proliferation and invasion of non-small cell lung cancer by regulating FGF2. Oncol Lett. 2019;17(3):3425–3431. doi:10.3892/ol.2019.9954
31. Yan Y, Peng Y, Ou YZ, et al. microRNA-497-5p targeted SOX9 to inhibit proliferation, migration and invasion of glioma cells. Int J Clin Exp Pathol. 2016;9(8):8027–8036.
32. Feng L, Cheng K, Zang R, et al. miR-497-5p inhibits gastric cancer cell proliferation and growth through targeting PDK3. Biosci Rep. 2019;39(9). doi:10.1042/BSR20190654.
33. Liu Z, Yao Y, Huang S, et al. LINC00662 promotes gastric cancer cell growth by modulating the Hippo-YAP1 pathway. Biochem Biophys Res Commun. 2018;505(3):843–849. doi:10.1016/j.bbrc.2018.09.191
34. Galluzzi L, Pietrocola F, Bravo-San Pedro JM, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34(7):856–880. doi:10.15252/embj.201490784
35. Dower CM, Wills CA, Frisch SM, Wang HG. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy. 2018;14(7):1110–1128. doi:10.1080/15548627.2018.1450020
36. Chi KH, Wang YS, Huang YC, et al. Simultaneous activation and inhibition of autophagy sensitizes cancer cells to chemotherapy. Oncotarget. 2016;7(36):58075–58088. doi:10.18632/oncotarget.10873
37. Park JM, Jung CH, Seo M, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12(3):547–564. doi:10.1080/15548627.2016.1140293
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.