Back to Journals » Cancer Management and Research » Volume 12
LINC01535 Promotes the Development of Osteosarcoma Through Modulating miR-214-3p/KCNC4 Axis
Authors Yao X, Wu L, Gu Z, Li J
Received 27 September 2019
Accepted for publication 9 May 2020
Published 8 July 2020 Volume 2020:12 Pages 5575—5585
DOI https://doi.org/10.2147/CMAR.S232757
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Xiaoke Yao,1,* Lingna Wu,2,* Zuchao Gu,1 Jianhua Li1
1Department of Orthopedics, Chengdu First People’s Hospital, Chengdu 610041, Sichuan, People’s Republic of China; 2Intensive Care Unit, Chengdu First People’s Hospital, Chengdu, Sichuan, 610041, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianhua Li
Department of Orthopedics, Chengdu First People’s Hospital, Chengdu 610041, Sichuan, People’s Republic of China
Email [email protected]
Background: Osteosarcoma (OS) is the most common primary bone tumor in group of children and adolescents. Increasing studies showed that long non-coding RNAs (lncRNAs) exerted important functions in the development of tumors, including OS. LINC01535 is an lncRNA which has been studied in cervical cancer but not in OS.
Aim of the Study: This study was aimed to explore the biological function and mechanism of LINC01535 in OS.
Methods: LINC01535 expression was detected by qRT-PCR. Colony formation assay, EdU assay and CCK-8 assay were applied to check cell proliferation ability in OS. Flow cytometry analysis was conducted to measure cell apoptosis capacity. Wound healing assay and transwell assay were performed to assess cell migration and invasion. Luciferase reporter assay and RNA pull-down assay were carried out to verify the molecular mechanism.
Results: The high expression of LINC01535 was presented in OS tissues and cell lines compared with adjacent normal tissues and human osteoblasts. Moreover, OS patients with high LINC01535 expression exhibited poor prognosis. Loss-of-function assay revealed that silenced LINC01535 significantly attenuated cell proliferation, migration and invasion, and enhanced cell apoptosis in OS. Through mechanistic exploration, we found that LINC01535 interacted with miR-214-3p, and KCNC4 was validated to be a target gene of miR-214-3p. The levels of KCNC4 mRNA and protein were positively modulated by LINC01535 and reversely mediated by miR-214-3p. Based on rescue experiments, KCNC4 overexpression reserved the suppressive function of silenced LINC01535 on OS cell growth, migration and invasion.
Conclusion: LINC01535, miR-214-3p and KCNC4 constituted an effective axis that exerted a pregnant regulation in OS development, which is a quite meaningful discovery for exploring potential therapeutic methods for OS patients.
Keywords: LINC01535, miR-214-3p, KCNC4, osteosarcoma
Introduction
Osteosarcoma (OS) is the most primary malignant bone cancer occurring in children and adolescents.1 Due to great advancements in the treatment strategies, such as radiotherapy, adjuvant chemotherapy and surgery (wider range of tumor resection), the overall survival of most OS patients has been significantly improved.2 However, the survival of patients with local recurrence or distant metastasis keeps poor.2 For exploring specific biomarkers and therapeutic targets, it is quite necessary to understand the molecular mechanism underlying OS development.
Long noncoding RNAs (lncRNAs) are a class of RNA transcripts with a length of over 200 nucleotides and without ability of protein coding.3–6 Increasing evidence showed that lncRNA was abnormally expressed in many human tumors.7,8 Recently, certain lncRNAs have been reported to play a regulatory role in the proliferation, apoptosis, migration and invasion of various tumor cells.9–12 For example, lncRNA HOTTIP is overexpressed and exacerbates cell proliferation and invasion in colorectal cancer.13 LncRNA UCA1 hastens cell cycle and migration and predicts a poor prognosis in gastric cancer.14 In mechanism, competing endogenous RNA (ceRNA) hypothesis was reported as a classical pattern to regulate gene expression. The ceRNA network suggested that lncRNA could function as microRNA (miRNA) sponge via competitively binding miRNAs to abolish the inhibitive function of miRNAs in specific target messenger-RNAs (mRNAs).15,16 Increasing studies reported that lncRNA, localizing in cytoplasmic, acted as ceRNA to affect tumorigenesis and progression in cancers.17,18 As reported, lncRNA UCA1 serves as a ceRNA in prostate cancer and promotes cancer progression via sponging miR143 and up-regulating MYO6.19 LncRNA NR2F1-AS1 acts as a ceRNA in OS and enhances cell growth by targeting miR-483-3p/FOXA1 axis.20 Previously, LINC01535 functioned as a ceRNA in cervical cancer and accelerated occurrence and development.21 However, the function and molecular mechanism of LINC01535 in OS have not been studied.
In this study, we confirmed that expression of LINC01535 presented a significant up-regulation in OS tissues and cells and promoted the development of OS. In addition, LINC01535 sponged miR-214-3p to up-regulate KCNC4. This finding might provide a theoretic basis for exploring treatment in OS.
Materials and Methods
Human Tissue Samples
Fifty paired OS tissues and adjacent normal tissues were obtained from 50 patients at Chengdu First People’s Hospital. The fresh tissues were snap-frozen in liquid nitrogen and stored at −80°C until use. Patients did not accept any therapy before operation and they all signed written informed consent. Ethical approval was obtained from Chengdu First People’s Hospital.
Cell Culture
Human OS cell lines (MG63, Saos2, HOS, U2OS) and human osteoblasts (hFOB1.19) were purchased from Chinese Academy of Sciences (Beijing, China). Cells were all incubated in DMEM (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS) (Gibco) plus 100mg/mL penicillin and streptomycin (Invitrogen, Carlsbad, CA, USA). Cells were developed in a humidified atmosphere with 5% CO2 at 37°C and the medium was replaced every 3 days.
Quantitative Real-Time PCR (qRT-PCR)
Treated cells were lysed by the use of Trizol reagent (Invitrogen), and the total RNA was extracted and then reversely transcribed into cDNA using a Reverse Transcription Kit (Invitrogen). qRT-PCR was processed by using SYBR Green RT-PCR Kit (Invitrogen) on an Applied Biosystems 7300 (Invitrogen) to detect the expression levels of RNA. The relative expression of indicated gene was calculated through 2−ΔΔCt method and the level of GAPDH/U6 was measured as an internal reference.
Cell Transfection
The short hairpin RNAs (shRNAs) for LINC01535 (sh-LINC01535-1/2) and their negative control (sh-NC) were purchased from Genechem (Shanghai, China). The miR-214-3p mimics and NC mimics, as well as the pcDNA3.1 vector expressing KCNC4 and empty vector, were obtained from Genechem. Cells were routinely transfected by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) for 48 h.
Cell Viability Assay
Briefly, cells were placed in 96-well plates under a density of 1 × 103 cells/well and cultured for indicated time points. Then, the solution of cell counting kit-8 (CCK-8) was added into each well and the absorbance at 450 nm was measured using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Colony Formation Assay
U2OS and Saos2 cells were seeded in 6-well plates after transfection. The cells were then fixed with 4% paraformaldehyde (Solarbio, Beijing, China) for 10 min and stained with 0.4% crystal violet (Solarbio). Finally, the number of visible colonies was determined.
Western Blot
The cell protein samples were extracted in 6-well plates using RIPA lysis buffer with protease inhibitor, then quantitated by BCA kit (Thermo Fisher Scientific), diluted in loading buffer to the same concentration, denatured at 95°C. Then, 20 μg proteins was subjected to SDS-PAGE for 2h and then transferred to PVDF membranes. After being blocked with skim milk, the membranes were co-cultured with primary antibodies against anti-KCNC4 (ab93605, Cambridge, Abcam), anti-MMP2 (ab37150, Abcam), anti-MMP7 (ab5706, Abcam), anti-MMP9 (ab38898, Abcam), anti-Slug (ab27568, Abcam), anti-Twist (ab50887, Abcam) and anti-GAPDH (ab8245, Abcam). Chemiluminescence detection system (GE Healthcare, Chicago, IL, USA) was employed to measure the protein.
5-Ethynyl-2′-Deoxyuridine (EdU) Labeling
Cells were seeded into 96-well plates and 100 μL of EdU reagent was added and cultured for 2 h. After washing with PBS (Solarbio), the cells were dyed with Apollo and then washed with methanol (Solarbio). Later, the cells were fixed, discolored and permeated. Subsequently, the cells were cultivated with 100 μL of DAPI reaction solution (Sigma-Aldrich Chemical Company, St Louis, MO, USA) followed by viability determination via a fluorescent microscope (Thermo Fisher Scientific).
Cell Apoptosis Analysis
Cultured Saos2 and U2OS cells were collected after transfection and rinsed in chilled PBS, followed by double staining with Annexin V fluorescein isothiocyanate (FITC)/propidium iodide (PI) detection kit (Invitrogen) in the dark for 15 min. At length, the rates of apoptotic cells were detected by flow cytometer (Beckman Coulter, Brea, CA, USA).
Wound Healing Assay
5 × 104 transfected cells were seeded in 96-well plates and cultured all night to allow cell lines to adhere. The wounds were scratched by sterile pipette tip when cells reached confluence. After washing in PBS, cells were observed at 0 and 24 h time interval.
Transwell Invasion Assay
Cell invasion in this study was examined by Matrigel-coated transwell chamber (Corning Co, Corning, NY), following the user guide. A total of 5 × 103 transfected cell lines were added to the upper chamber with serum-free medium and lower chamber was added with the 100% complete culture medium. Cells in the lower chambers were fixed by 4% PFA after 24 h of incubation, and the invaded cells were stained by crystal violet solution. Five random fields were selected for counting under microscope (magnification, ×200).
In vivo Assay
The 6-week-old BALB/C athymic nude mice were procured from the National Laboratory Animal Center (Beijing, China). Animal studies have obtained the approval from the Animal Research Ethics Committee of Chengdu First People’s Hospital. In vivo assay was undertaken via subcutaneous injection of 5 × 106 transfected cells into mice, the tumor volume was monitored every 4 days. After 28 days, mice were sacrificed and tumors were excised for subsequent analysis.
Immunohistochemistry (IHC)
The tissue samples collected from in vivo assay were prepared to fix with 4% PFA, subsequently dehydrated and embedded in paraffin. IHC analysis was performed in the consecutive 4 μm thick sections using specific primary antibodies and secondary antibodies.
Luciferase Reporter Analysis
The sequences for wild-type LINC01535/KCNC4 or mutant LINC01535/KCNC4 containing the putative binding sites of miR-214-3p were cloned into the pmirGLO dual-luciferase vector (Promega, Madison, WI, USA). The vectors were co-transfected with miR-214-3p mimics and NC mimics into U2OS and Saos2 cells with Lipofectamine 2000 (Invitrogen). Luciferase activities were tested with Dual-Luciferase Reporter Assay System (Promega).
RNA Pull-Down Assay
The RNA interaction in this study was examined following the instruction of Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA). Biotin-labeled RNAs (Bio-miR-214-3p-WT/Bio-miR-214-3p-Mut) and control (Bio-miR-NC) were acquired using the Biotin RNA Labeling Mix (Roche, Mannheim, Germany), followed by treating with RNase-free DNase I (Roche) and purified by the RNeasy Mini Kit (Qiagen, Hilden, Germany) as per the specification. Cell protein extracts were prepared by RIPA lysis buffer, and then mixed with biotinylated probes and 50 μL of streptavidin agarose magnetic beads. The RNA-protein binding mixture was finally analyzed by qRT-PCR.
RNA Immunoprecipitation (RIP) Assay
Ago2-RIP assay was performed through Magna RIP™ RNA Binding Protein Immunoprecipitation Kit (Millipore) based on the guides. When the cell fusion rate was 80%-90%, cells were reaped and lysed in RIP lysis buffer (Invitrogen) supplemented with magnetic bead conjugated with antibodies of anti-Ago2 (Abcam) or anti-IgG (Abcam) overnight at 4°C. At last, the purified immunoprecipitated RNA was detected by qRT-PCR analysis.
Subcellular Fractionation Assay
RNA separated from nuclear or cytoplasm fraction by the use of the Nuclear/Cytosol Fractionation Kit (Biovision, San Francisco Bay, CA, USA) was detected with qRT‐PCR analysis. U6 or GAPDH was seen as the nuclear or cytoplasmic control, separately.
Fluorescence in situ Hybridization (FISH) Assay
FISH kit (Roche Diagnostics GmbH, Mannheim, Germany) was applied to detect the subcellular localization of LINC01535. The cells were cultivated with hybridization solution containing LINC01535 specific probe (Sigma-Aldrich). DAPI (Sigma-Aldrich) was used to stain the cell nucleus. The images were captured under a microscope.
Statistical Analysis
Data were denoted as means ± SD. Statistical analysis was conducted using GraphPad Prism 5 software (GraphPad Software, San Diego, CA). Significance of the variance between two or multiple groups was evaluated by Student’s t-test or ANOVA. Kaplan–Meier method was used to analyze the overall survival. P < 0.05 was seen as statistical significance in general. All experiments were performed in triplicate.
Results
The High Expression of LINC01535 Accelerates Proliferation, Migration, Invasion and Decelerates Apoptosis of OS Cells
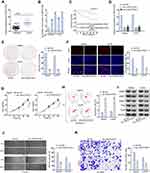
First of all, we examined LINC01535 expression in OS tissues and cell lines (MG63, Saos2, HOS and U2OS) by qRT-PCR. The adjacent normal tissues and human normal osteoblasts (hFOB1.19) were taken as control, separately. As depicted in Figure 1A, the expression of LINC01535 was significantly higher in OS tissues compared with adjacent normal tissues. Similarly, LINC01535 expression in OS cell lines was also higher than that in hFOB1.19 cell line (Figure 1B). Due to relatively higher expression of LINC01535 in Saos2 and U2OS cells, the two cells were selected for the following experiments. Then, we analyzed the prognostic potential of LINC01535 in OS patients by Kaplan–Meier surviving curves. It was displayed that high expression level of LINC01535 was closely associated with the low overall survival rate of OS patients (Figure 1C). Thus, we investigated the functional role of LINC01535 in OS cellular progression through loss-of-function assay. Firstly, we tested the interference efficiency of LINC01535 by transfecting with sh-LINC01535-1 and sh-LINC01535-2 in Saos2 and U2OS cell lines. The results showed that LINC01535 expression was obviously decreased after sh-LINC01535-1/2 transfection (Figure 1D). The interference efficiency was more effective in sh-LINC01535-1 group; therefore, we selected it for the subsequent assays. Then, we implemented colony formation assay, EdU assay and CCK-8 assay to assess the effect of silenced LINC01535-1 on OS cell proliferation. These results all indicated the descending proliferative ability of Saos2 and U2OS cells after LINC01535 knockdown (Figure 1E–G). In addition, we detected the capacity of cell apoptosis by flow cytometry analysis in Saos2 and U2OS cells. The consequence indicated that cell apoptotic ability was remarkably enhanced after silencing LINC01535 (Figure 1H). Later, the levels of cell migration-associated proteins (MMP2, MMP7, MMP9, Slug and Twist) were tested by Western blot to determine cell migration upon LINC01535 knockdown. As the result demonstrated, cell migration-associated protein levels were all decreased by LINC01535 deficiency (Figure 1I). Furthermore, wound healing assay further revealed that LINC01535 silencing curbed the migratory ability of Saos2 and U2OS cells (Figure 1J). In transwell assay, we observed that the invasion was also hindered in Saos2 and U2OS cells transfected with sh-LINC01535-1 (Figure 1K). Finally, we conducted in vivo assay to further verify the results obtained from in vitro assay. As we observed, the tumors in sh-LINC01535-1 group were smaller than those in sh-NC group (Figure S1A). Consistently, the tumor growth curve was also lower in sh-LINC01535-1 group compared with sh-NC group (Figure S1B). Besides, tumor weight of LINC01535 knockdown group was obviously decreased with comparison of control group (Figure S1C). In excised tumors, LINC01535 expression was also lessened in sh-LINC01535 group (Figure S1D). Moreover, IHC assay confirmed that LINC01535 deficiency could reduce the expressions of Ki-67 and PCNA (proliferation markers) (Figure S1E). These data suggested that LINC01535 acted as an oncogene in OS.
LINC01535 Interacts with miR-214-3p
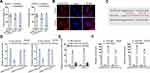
Primarily, to confirm the localization of LINC01535, we made subcellular fractionation assay in Saos2 and U2OS cell lines. The results indicated that LINC01535 was mostly distributed in cytoplasm (Figure 2A). Additionally, FISH experiment further validated the cytoplasmic location of LINC01535 in OS (Figure 2B). Above results indicated that LINC01535 could play a role at post-transcriptional level, which provided evidence for ceRNA hypothesis. To find out the specific miRNA, we employed starBase (http://starbase.sysu.edu.cn/index.php) and found eleven miRNAs that could bind to LINC01535. To further filtrate, we checked expressions of predicted eleven miRNAs in normal human osteoblasts and OS cells. The results revealed that miR-214-3p expression was significantly down-regulated in OS cells, while other miRNAs did not exhibit expression difference (Figure S2A–K). Most interestingly, we also found the binding site between LINC01535 and miR-214-3p (Figure 2C). In addition, we conducted luciferase reporter assay via co-transfecting LINC01535-WT/Mut and miR-214-3p mimics or NC mimics into Saos2 and U2OS cells. The results presented that the luciferase activity of LINC01535-WT was eminently declined by miR-214-3p overexpression, whereas no change was observed in LINC01535-Mut group (Figure 2D). Moreover, to certify the direct interaction between LINC01535 and miR-214-3p, RIP assay and RNA pull-down assay were carried out. According to the results of RNA pull-down experiment, we discovered that the enrichment of LINC01535 was significantly elevated by biotinylated miR-214-3p-WT instead of biotinylated miR-214-3p-Mut (Figure 2E). Besides that, LINC01535 and miR-214-3p were both enriched in Ago2-conjugated beads (Figure 2F). Lastly, we tested miR-214-3p expression in sh-LINC01535-1 transfected cells. The results displayed that LINC01535 silencing could not affect miR-214-3p expression (Figure S2L). These results manifested LINC01535 sponged miR-214-3p through ceRNA network rather than directly regulated miR-214-3p.
LINC01535 Positively Modulates KCNC4 Expression via Sponging miR-214-3p
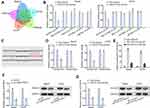
Subsequently, we explored the target genes of miR-214-3p and screened out five mRNAs, which could bind to miR-214-3p by using starBase (Figure 3A). Additionally, we performed luciferase reporter assay to estimate the luciferase activity of KCNC4 3ʹUTR, TBCID10B 3ʹUTR, SNX12 3ʹUTR, SPTBN2 3ʹUTR and REPIN1 3ʹUTR after transfecting miR-214-3p mimics. It was shown that KCNC4 3ʹUTR activity was prominently decreased by miR-214-3p mimics, while there was no alteration in other groups (Figure 3B). Hence, KCNC4 was predicted as a target gene of miR-214-3p. Further, we obtained potential binding site between KCNC4 and miR-214-3p from starBase (Figure 3C). And we found the luciferase activity of KCNC4-WT reporter was clearly decreased whereas there was no difference in KCNC4-Mut group with transfection of miR-214-3p mimics (Figure 3D). This consequence certified the interaction between miR-214-3p and KCNC4. Then, RNA pull-down assay was performed in Saos2 and U2OS cells. The results indicated that KCNC4 could be pulled down by biotinylated miR-214-3p-WT rather than biotinylated miR-214-3p-Mut (Figure 3E). Furthermore, we determined the KCNC4 mRNA expression and protein level after transfecting with sh-LINC01535-1 or miR-214-3p mimics. The results manifested that the expressions of KCNC4 mRNA and protein were both attenuated by sh-LINC01535-1 transfection (Figure 3F). Simultaneously, the reduced KCNC4 mRNA and protein levels were observed in miR-214-3p mimics group (Figure 3G). Taken together, LINC01535 positively modulated KCNC4 expression via sponging miR-214-3p.
LINC01535 Facilitates Cell Growth, Migration and Invasion in OS by Regulating KCNC4
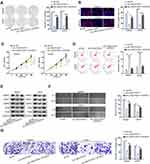
Finally, we carried out a battery of functional rescue experiments to explore whether LINC01535 modulated OS progression by regulating KCNC4. Colony formation assay, EdU assay and CCK-8 assay were conducted to detect cell proliferation ability. The results displayed that cell proliferation was attenuated after silencing LINC01535, while this phenomenon was recovered by overexpressing KCNC4 (Figure 4A–C). In addition, flow cytometry analysis unveiled that cell apoptosis elevated by LINC01535 interference was subsequently restored by KCNC4 overexpression (Figure 4D). Through Western blot analysis, we observed levels of migration-related proteins were descending remarkably by reducing LINC01535. However, the protein levels were ascending upon KCNC4 overexpression (Figure 4E). As demonstrated in wound healing assay, KCNC4 overexpression recovered the inhibitory role of LINC01535 depletion in the migratory capacity of OS cells (Figure 4F). Finally, transwell assay confirmed the restorative effect of KCNC4 up-regulation on the suppressive invasion of sh-LINC01535-1 transfected cells (Figure 4G). Above experimental data demonstrated that LINC01535 regulated KCNC4 to promote cell growth, migration and invasion in OS.
Discussion
OS is one of the leading causes of cancer-related death in children and adolescents. It is difficult for patients to make meaningful progress in survival rate because of distant metastasis and large tumor heterogeneity.22 However, it is still necessary to explore effective biomarkers and targeted therapies for OS patients. LncRNAs have been manifested as regulators in various tumor processes.23–25 In the meantime, dysregulation of lncRNA was associated with the development of disease.26 Previously, LINC01535 has been certified to facilitate cell development in cervical cancer.21 Thus, we wondered whether LINC01535 was an oncogene and involved in the pathogenesis of OS.
Based on the previous study, we examined the expression of LINC01535 in tumor tissues and cells of OS and found that its expression level was up-regulated in OS. Meanwhile, the survival rate of patients with high LINC01535 expression level was significantly lower than who with low LINC01535 expression level, suggesting that up-regulated LINC01535 resulted in poor prognosis. Besides, LINC01535 knockdown overtly suppressed OS cell proliferation, migration and invasion, and promoted OS cell apoptosis in vitro. In vivo assay uncovered that silenced LINC01535 hampered tumor growth. Therefore, we had reason to believe that LINC01535 also acted as an oncogene in OS.
In general, accumulating studies showed that lncRNAs could function as ceRNA to sponge miRNA.27,28 MiRNAs also act as tumor regulator in the biological process and they are vital RNAs with a length of 21–25 nucleotides.29,30 For example, down-regulated miR-193a-5p induces docetaxel chemosensitivity in prostate cancer.31 In melanoma, miR-204-5p and miR-211-5p promote the resistance of BRAF inhibitor.32 Besides, miR-361-5p targets FGFR1 and MMP-1 in breast cancer and restrains cell proliferation and invasion.33 According to relevant reports, miR-214-3p presented down-regulated expression in tumors and affected tumor occurrence and development.34–37 Although LINC01535 has been reported as a ceRNA in cervical cancer, but its mechanism in OS has not been investigated.21 Here, we discovered that LINC01535 was cytoplasmic RNA and sponged miR-214-3p in OS. Subsequently, KCNC4 was identified as a downstream gene of miR-214-3p. KCNC4 was reported to be highly expressed in head and neck squamous cell carcinoma and aggravate cancer malignancy.38 In our study, the expression of KCNC4 was positively mediated by LINC01535 and reservedly modulated by miR-214-3p. Functional rescue assays suggested that up-regulated KCNC4 counteracted LINC01535 deficiency-mediated suppression on OS cell growth, migration and invasion.
It was concluded that LINC01535 enhanced OS cellular progression via secluding miR-214-3p and up-regulating KCNC4, which constituting an effective axis of LINC01535/miR-214-3p/KCNC4 in OS. Simultaneously, this network might provide theoretical basis for the exploration of prevention and remedy treatment of OS.
Data Sharing Statement
Not applicable.
Ethics Approval and Informed Consent
Patients did not accept any therapy before operation and they all signed written informed consent. Ethical approval was obtained from Chengdu First People’s Hospital. Animal studies have obtained the approval from the Animal Research Ethics Committee of Chengdu First People’s Hospital.
Consent for Publication
All authors have read and approved the final manuscript for publication.
Acknowledgments
We appreciate all participants who provide technical supports for our research. Xiaoke Yao and Lingna Wu are co-first authors for this study.
Author Contributions
This manuscript was written by Xiaoke Yao, Lingna Wu, Zuchao Gu and Jianhua Li, who made significant contributions in conception and design, data acquisition, data analysis and interpretation, writing articles or critical modification of important intellectual content. All authors gave final approval of the version to be published, agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors declare no conflicts of interest in this study.
References
1. Xie L, Yao Z, Zhang Y, et al. Deep RNA sequencing reveals the dynamic regulation of miRNA, lncRNAs, and mRNAs in osteosarcoma tumorigenesis and pulmonary metastasis. Cell Death Dis. 2018;9(7):772. doi:10.1038/s41419-018-0813-5
2. Liu K, Hou Y, Liu Y, Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J Biomed Sci. 2017;24(1):46. doi:10.1186/s12929-017-0353-9
3. Lin A, Hu Q, Li C, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19(3):238–251. doi:10.1038/ncb3473
4. Yuan JH, Liu XN, Wang TT, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19(7):820–832. doi:10.1038/ncb3538
5. Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. 2018;19(9):535–548. doi:10.1038/s41576-018-0017-y
6. Pop S, Enciu AM, Necula LG, Tanase C. Long non-coding RNAs in brain tumours: focus on recent epigenetic findings in glioma. J Cell Mol Med. 2018;22(10):4597–4610. doi:10.1111/jcmm.13781
7. Tang GH, Chen X, Ding JC, et al. LncRNA LUCRC regulates colorectal cancer cell growth and tumorigenesis by targeting endoplasmic reticulum stress response. Front Genet. 2019;10:1409. doi:10.3389/fgene.2019.01409
8. Dou X, Zhou Q, Wen M, et al. Long noncoding RNA FOXD2-AS1 promotes the malignancy of cervical cancer by sponging MicroRNA-760 and upregulating hepatoma-derived growth factor. Front Pharmacol. 2019;10:1700. doi:10.3389/fphar.2019.01700
9. Chen Z, Xu D, Zhang T. Inhibition of proliferation and invasion of hepatocellular carcinoma cells by lncRNA-ASLNC02525 silencing and the mechanism. Int J Oncol. 2017;51(3):851–858. doi:10.3892/ijo.2017.4069
10. Mao Z, Li H, Du B, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37(6). doi:10.1042/BSR20171070.
11. Min L, Hong S, Duan H, et al. Antidifferentiation noncoding RNA regulates the proliferation of osteosarcoma cells. Cancer Biother Radiopharm. 2016;31(2):52–57. doi:10.1089/cbr.2015.1888
12. Wang H, Yu Y, Fan S, Luo L. Knockdown of long non-coding RNA NEAT1 inhibits proliferation and invasion and induces apoptosis of osteosarcoma by inhibiting miR-194 expression. Yonsei Med J. 2017;58(6):1092–1100. doi:10.3349/ymj.2017.58.6.1092
13. Liu T, Wang H, Yu H, et al. The long non-coding RNA HOTTIP is highly expressed in colorectal cancer and enhances cell proliferation and invasion. Mol Ther Nucleic Acids. 2019;19:612–618. doi:10.1016/j.omtn.2019.12.008
14. He X, Wang J, Chen J, et al. lncRNA UCA1 predicts a poor prognosis and regulates cell proliferation and migration by repressing p21 and SPRY1 expression in GC. Mol Ther Nucleic Acids. 2019;18:605–616. doi:10.1016/j.omtn.2019.09.024
15. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
16. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi:10.1158/2159-8290.CD-13-0202
17. Tay Y, Kats L, Salmena L, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–357. doi:10.1016/j.cell.2011.09.029
18. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
19. Yu Y, Gao F, He Q, Li G, Ding G. lncRNA UCA1 functions as a ceRNA to promote prostate cancer progression via sponging miR143. Mol Ther Nucleic Acids. 2019;19:751–758. doi:10.1016/j.omtn.2019.11.021
20. Li S, Zheng K, Pei Y, Wang W, Zhang X. Long noncoding RNA NR2F1-AS1 enhances the malignant properties of osteosarcoma by increasing forkhead box A1 expression via sponging of microRNA-483-3p. Aging. 2019;11(23):11609–11623. doi:10.18632/aging.102563
21. Song H, Liu Y, Jin X, et al. Long non-coding RNA LINC01535 promotes cervical cancer progression via targeting the miR-214/EZH2 feedback loop. J Cell Mol Med. 2019;23(9):6098–6111. doi:10.1111/jcmm.14476
22. Botter SM, Neri D, Fuchs B. Recent advances in osteosarcoma. Curr Opin Pharmacol. 2014;16:15–23. doi:10.1016/j.coph.2014.02.002
23. Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, Kashani HH, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16(1):107. doi:10.1186/s12943-017-0671-2
24. Yang Y, Junjie P, Sanjun C, Ma Y. Long non-coding RNAs in colorectal cancer: progression and future directions. J Cancer. 2017;8(16):3212–3225. doi:10.7150/jca.19794
25. Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283(20):3739–3754. doi:10.1111/febs.13839
26. Liu T, Yu T, Hu H, He K. Knockdown of the long non-coding RNA HOTTIP inhibits colorectal cancer cell proliferation and migration and induces apoptosis by targeting SGK1. Biomed Pharmacother. 2018;98:286–296. doi:10.1016/j.biopha.2017.12.064
27. Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2019.
28. Kang M, Shi J, Li B, Luo M, Xu S, Liu X. LncRNA DGCR5 regulates the non-small cell lung cancer cell growth, migration, and invasion through regulating miR-211-5p/EPHB6 axis. BioFactors (Oxford, England). 2019;45(5):788–794. doi:10.1002/biof.1539
29. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
30. Mohr S, Doebele C, Comoglio F, et al. Hoxa9 and Meis1 cooperatively induce addiction to Syk signaling by suppressing miR-146a in acute myeloid leukemia. Cancer Cell. 2017;31(4):549–562.e511. doi:10.1016/j.ccell.2017.03.001
31. Yang Z, Chen JS, Wen JK, et al. Silencing of miR-193a-5p increases the chemosensitivity of prostate cancer cells to docetaxel. J Exp Clin Cancer Res CR. 2017;36(1):178. doi:10.1186/s13046-017-0649-3
32. Diaz-Martinez M, Benito-Jardon L, Alonso L, Koetz-Ploch L, Hernando E, Teixido J. miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res. 2018;78(4):1017–1030. doi:10.1158/0008-5472.CAN-17-1318
33. Ma F, Zhang L, Ma L, Zhang Y, Zhang J, Guo B. MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. J Exp Clin Cancer Res CR. 2017;36(1):158. doi:10.1186/s13046-017-0630-1
34. Li AH, Zhang HH. Overexpression of lncRNA MNX1-AS1 is associated with poor clinical outcome in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21(24):5618–5623. doi:10.26355/eurrev_201712_14003
35. Wang X, Li H, Shi J. LncRNA HOXA11-AS promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppression of miR-214-3p expression. Biomed Res Int. 2019;2019:8645153.
36. Peng W, Zhu S, Chen J, Wang J, Rong Q, Chen S. Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed Pharmacother. 2019;109:1709–1717. doi:10.1016/j.biopha.2018.10.159
37. Zhan M, He K, Xiao J, et al. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018;22(8):3758–3767. doi:10.1111/jcmm.13633
38. Menendez ST, Rodrigo JP, Allonca E, et al. Expression and clinical significance of the Kv3.4 potassium channel subunit in the development and progression of head and neck squamous cell carcinomas. J Pathol. 2010;221(4):402–410. doi:10.1002/path.2722
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.