Back to Journals » OncoTargets and Therapy » Volume 13
LINC01436 Promotes the Progression of Gastric Cancer via Regulating miR-513a-5p/APE1 Axis
Received 10 April 2020
Accepted for publication 19 September 2020
Published 19 October 2020 Volume 2020:13 Pages 10607—10619
DOI https://doi.org/10.2147/OTT.S257747
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
This paper has been retracted.
Ming-dian Lu, Dong Liu, Yong-xiang Li
Department of Gastrointestinal Surgery and General Surgery, First Affiliated Hospital of Anhui Medical University, Hefei 230022, Anhui, People’s Republic of China
Correspondence: Yong-xiang Li
Department of Gastrointestinal Surgery and General Surgery, First Affiliated Hospital of Anhui Medical University, Jixi Avenue No. 218, Hefei 230022, Anhui Province, People’s Republic of China
Tel +86 551-62922114
Email [email protected]
Background: Gastric cancer (GC) is one of the deadliest cancer worldwide. Multiple long non-coding RNAs (lncRNAs) are recently identified as crucial oncogenic factors or tumor suppressors in GC. In this study, we aimed to probe into the effect of LINC01436 on GC progression.
Methods: LINC01436 and miR-513a-5p expressions in GC tissue samples were measured using quantitative real-time polymerase chain reaction (qRT-PCR). Western blot was used to detect the expression of apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1) expression. Human GC cell lines AGS and BGC-823 were employed to investigate the function and mechanism of LINC01436 in GC. Cell counting kit-8 (CCK-8) assay was used to assess the effect of LINC01436 on proliferation. Flow cytometry was utilized to explore the effect of LINC01436 on apoptosis, and Transwell assay was conducted to detect the effect of LINC01436 on the migration and invasion. Colony formation assay was performed to evaluate the effect of LINC01436 on radioresistance of GC cells. Furthermore, luciferase reporter assay and RNA immunoprecipitation assay were conducted to confirm the binding relationship between miR-513a-5p and LINC01436. Additionally, Western blot was used to study the regulatory function of LINC01436 and miR-513a-5p on APE1.
Results: LINC01436 expression of GC clinical samples was remarkably increased and LINC01436 was correlated with unfavorable pathological indexes. LINC01436 high expression was associated with shorter overall survival time. Its overexpression observably promoted the proliferation, metastasis and radioresistance of GC cells, and its knockdown suppressed the malignant phenotypes of GC cells. LINC01436 overexpression markedly reduced the miR-513a-5p expression via sponging it and enhanced the APE1 expression. MiR-513a-5p overexpression or APE1 knockdown reversed the effects of LINC01436 on GC cells.
Conclusion: LINC01436 is a molecular sponge of tumor suppressor miR-513a-5p, which indirectly enhances the APE1 expression and functions as the oncogenic lncRNA in GC.
Keywords: gastric cancer, LINC01436, miR-513a-5p, APE1, ceRNA
Background
Gastric cancer (GC) is one of the most common cancers in the world, whose mortality rate ranks the second among cancers.1 Due to the limited early diagnosis technique of GC, high recurrence rate and high metastasis rate, the prognosis of patients with GC is poor.2 Therefore, to probe the potential biological mechanisms of GC is of great significance for the prevention and treatment of this disease.
Accumulating studies find that long non-coding RNAs (lncRNAs) are abnormally expressed in a variety of malignancies and can affect the progression of multiple cancers including GC.3,4 For instance, abnormal upregulation of lncRNA PANDAR expression is a poor prognostic factor for predicting cervical cancer;5 down-regulation of lncRNA MALAT1 expression suppresses the metastasis of breast cancer.6 It is reported that LINC01436 accelerates the growth and metastasis of NSCLC cells, implying the oncogenic role of LINC01436 in cancer biology.7 However, the specific regulatory mechanism of LINC01436 in GC is much less explored.
MicroRNAs (miRNAs) are a class of non-coding RNA (ncRNA) molecules of 21 to 25 nt in length, which are highly conserved and cannot encode proteins, and they are involved in the regulation of tumor progression through post-transcriptional regulation of genes.8–11 MiR-513a-5p plays an essential role in modulating inflammatory response-mediated cell injury. It is reported that it is involved in TNF-α and LPS induced apoptosis via down-regulating the expression of X-linked inhibitors of apoptotic proteins in endothelial cells.12 The dysregulation of miR-513a-5p expression is validated to be closely linked to breast cancer risk.13 Nevertheless, the role of miR-513a-5p in GC progression remains to be studied.
Apurinic/apyrimidinic endodeoxyribonuclease 1/redox factor-1 (APE1/REF-1) is a multifunctional protein that is generally involved in repairing DNA injury through its endonuclease activity in base excision repair.14 It controls the activity of diverse transcription factors such as STAT3, NF-κB, AP-1, p53 and hypoxia-inducible factor 1α (HIF1α).15 APE1 also plays a key role in cancer progression. For instance, the APE1 expression in triple-negative breast cancer tissues is significantly higher than that in adjacent tissues, and APE1 is expected to be a prognostic factor for breast cancer.16,17 Particularly, APE1 is also involved in GC progression.18 Nonetheless, its upstream regulatory mechanism in GC remains to be further investigated.
LncRNA can function as a competitive endogenous RNA (ceRNA) of microRNA, thereby inhibiting gene expression and function. In our preliminary research, we noticed that miR-513a-5p had potential binding sites with both 3ʹUTR of APE1 mRNA and LINC01436. Hence, we hypothesized that in GC, LINC01436 high expression could facilitate the GC progression by inhibiting miR-513a-5p expression and indirectly facilitating APE1 expression.
Materials and Methods
Clinical Tissues Collection
All GC tissues were obtained from GC patients at First Affiliated Hospital of Anhui Medical University from May 2017 to March 2018. Forty cases of GC tissues were randomly selected in this study. The specimens in the control group were resected from paracancerous tissues of the same patient (at least 3 cm from the surgical margin), and no cancer cells were found in the pathological examination. All specimens were placed in liquid nitrogen at −196°C immediately after the removal. All patients involved gave informed consent and signed a written consent form. This study was ratified by the Ethics Review Committee of the First Affiliated Hospital of Anhui Medical University. All protocols were conducted in accordance with the principles of the Declaration of Helsinki.
Cell Lines and Cell Culture
The GC cell lines SGC7901, MKN45, BGC-823, AGS and human normal gastric epithelial cell GES-1 were purchased from the Cell Center of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured with RPMI 1640 medium (Thermo Fisher Scientific, Cambridge, MA, USA) containing 10% fetal bovine serum (FBS, Thermo Fisher Scientific, Cambridge, MA, USA) and 100 U/mL penicillin, and 100 U/mL streptomycin (Invitrogen, Carlsbad, CA, USA) in an incubator at 37°C in 5% CO2.
Cell Transfection
LINC01436 overexpression plasmid, LINC01436-siRNA (si-LINC01436), APE1 overexpression plasmid, APE1-siRNA (si-APE1), miR-513a-5p mimics, miR-513a-5p inhibitors and their corresponding controls were constructed by GenePharma (Shanghai, China). Lipofectamine® 3000 (Invitrogen, Carlsbad, CA, USA) was used to perform the transfection in line with the supplier’s instructions.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNA was extracted from tissues and cells with TRIzol reagent (Thermo Fisher Scientific, Shanghai, China), and then the RNA was reversely transcribed into cDNA using the TOYOBO Reverse Transcription Kit (TOYOBO, Osaka, Japan) following the manufacturer’s instructions. qRT-RCR was performed with SYBR premix EX TAQ II (Takara, Dalian, China) on an ABI 7500 Real-Time PCR System (Applied Biological System, CA, USA). GAPDH and U6 were used as the internal references of LINC01436, and APE1 and miR-513a-5p, respectively, and the relative expression levels were analyzed using the 2−ΔΔCt method. The primer sequences are shown in Table 1.
 |
Table 1 qRT-PCR Primer Sequences |
Western Blot
The cells were lysed with RIPA lysate (containing 1% PMSF), and the supernatant was collected after the high-speed centrifugation. Subsequently, the Bradford method was used for protein quantification. Then the sample was heated in a water bath at 100°C for 10 min. SDS-PAGE was used to separate the proteins and then the proteins were transferred onto polyvinylidene fluoride (PVDF) membranes. After that, the membrane was blocked at room temperature with 5% skimmed milk for 1 h, followed by being added with anti-Anti-APE1 antibody (ab76472) (1:1000; Abcam, Cambridge, UK) and Anti-β-actin antibody (ab115777) (1:1000; Abcam, Cambridge, UK) for incubation overnight at 4°C and then rinsed with TBST solution. Subsequently, the membrane was incubated with secondary antibodies (Beyotime, Shanghai, China) for 1 h at room temperature before being washed 3 times. At last, color rendering was performed using hypersensitive ECL (Biossci Biotechnology Co, Ltd., Wuhan, China).
Cell Counting Kit-8 (CCK-8) Assay
The AGS and BGC-823 cells in the logarithmic growth phase were prepared as a single cell suspension by 0.25% trypsinization. After counting the cells, the cells were seeded in 96-well plates at about 2000 cells per well and cultured at 37°C in 5% CO2. After the culture for 0, 1, 2, 3, and 4 d in an incubator, respectively, 10 μL of CCK-8 kit (Beyotime, Shanghai, China) was added to each well, and the incubation was continued for 1 h. Then the absorbance of each well was measured at a wavelength of 450 nm with a microplate reader.
Migration and Invasion Analysis
The migration and invasion were detected using the Transwell cell assays. In migration assay, 2×104 cells/well (without FBS) were seeded in the upper chamber of the Transwell chamber with an 8 μm pore size membrane (Corning, NY, USA), and the lower chamber was added with 500 μL of RPMI 1640 medium supplemented with 10% FBS. After being cultured for 24 h, the unmigrated cells were wiped off with a cotton swab. Cells passing through the membrane were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, photographed and counted after drying. In invasion assay, Matrigel (BD, San Jose, CA, USA) was used to coat the Transwell membrane before the cells were inoculated. The other procedures were the same as the migration assay.
Luciferase Report Analysis
The luciferase reporter assay was used to verify the targeting relationship between LINC01436 and miR-513a-5p. AGS cells were seeded at a density of 5 x 105 cells in a 24-well plate, and the luciferase reporter vectors containing wild type (WT) or mutant (Mut) LINC01436 sequence were cloned into pGL3 Basic vector (Promega, Madison, WI, USA), respectively, and then transfected into the cells. Following that, the miR-513a-5p mimics or negative control was co-transfected into the above cells, respectively. After 48 h of transfection, luciferase activity was determined using the dual-luciferase reporter assay system (Promega, Madison, WI, USA) in accordance with the manufacturer’s instructions.
RNA Immunoprecipitation (RIP) Assay
To pinpoint the relationship between LINC01436 and miR-513a-5p, RIP analysis was performed using the EZ-Magna RIP kit (Millipore, Billerica, MA, USA) complying with the manufacturer’s instructions. AGS cells were lysed using RIP lysis buffer containing protease inhibitors and RNase inhibitors, and then antibody against Ago2 or mouse IgG coupled with magnetic beads was incubated with the lysate for 1 h at room temperature. Subsequently, after removing the protein with proteinase K, the immunoprecipitated RNA was extracted for qRT-PCR analysis.
Flow Cytometry Analysis
The cells were trypsinized with trypsin and collected by centrifugation (1500 r/min, 3 min). Subsequently, the Annexin V‐FTIC/Propidium iodide (PI) Apoptosis Detection Kit (Aladdin BioReagent Co., Ltd., Shanghai, China) was used to detect cellular apoptosis. In brief, after the cells were washed twice with PBS, 400 μL of pre-cooled PBS was added to resuspend the cells, followed by the addition of 10 μL of Annexin V-FITC staining solution and 5 μL of PI staining solution. Thereafter, the cells were incubated at 4°C in the dark. After 30 min, the percentage of apoptotic cells was detected by a flow cytometer (Becton-Dickinson, Fullerton, CA, USA) immediately.
Colony Formation Analysis
First, 1000 cells in each group were seeded in a 35 mm culture dish. Then the cells were cultured for 24 h and then treated with different doses of irradiation (0 Gy, 4 Gy and 8 Gy). Subsequently, the cells were cultured for 2 weeks. Ultimately, the cells were stained with 0.1% crystal violet and washed with PBS, and the number of colonies was counted.
Mice Assay
The procedures in animal experiments were approved by Ethics Review Committee of the First Affiliated Hospital of Anhui Medical University. All the protocols of the animal experiments in this study complied with the United Kingdom Coordinating Committee on Cancer Research (UKCCCR) guidelines for welfare of animals in experimental neoplasia. Twelve six-week-old female BALB/c nude mice (SPF grade) were selected and randomly divided into two groups (si-NC group and si-LINC01436 group). 100 μL of AGS cell suspension (containing about 2×106 cells) was subcutaneously inoculated to the right (si-LINC01436) and left (si-NC) side of each mouse. The longest and shortest diameters of the tumor were measured once a week using a caliper until the tumor was removed after 5 weeks. The formula was adopted to calculate the tumor volume: volume = (length × width2 × 0.5).
Statistical Analysis
Statistical analysis was performed using SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA), and the measurement data were expressed as mean ± standard deviation (x ± s). Differences between the different groups were analyzed by t-test or one-way ANOVA. A significant statistical difference was considered at P < 0.05.
Results
LINC01436 Was Highly Expressed in GC Tissues and Cells
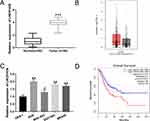
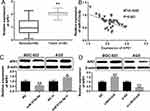
To investigate the expression characteristics of LINC01436 in GC, we examined the LINC01436 expression in 40 cases of cancerous tissues from GC patients and compared it with the LINC01436 expression in corresponding adjacent tissues. qRT-PCR analysis revealed that the LINC01436 expression in tumor tissues was observably higher than that in non-tumor tissues (Figure 1A). Consistently, GEPIA database (http://gepia.cancer-pku.cn/) unearthed that the LINC01436 expression was markedly up-regulated in GC tissues in comparison with the normal tissues (Figure 1B) (data were from The Cancer Genome Atlas, TCGA). Subsequently, the LINC01436 expression in each cell line was examined, the findings of which demonstrated that the LINC01436 expression was remarkably increased in the GC cell lines (AGS, BGC-823, SGC7901 and MKN45 cells) compared to in the normal cell line GES-1 (Figure 1C). To further fathom the clinical significance of LINC01436 expression in GC, we explored the association between the LINC01436 expression and the clinical features of GC patients. Forty patients were divided into LINC01436 high expression group and LINC01436 low expression group in accordance with the LINC01436 median expression. Our data demonstrated that LINC01436 high expression in GC tissues was significantly associated with T stage and differentiation status, but not with age, gender, tumor size, and lymphatic metastasis (Table 2). Importantly, TCGA data showed that the overall survival time of patients with high expression of LINC01436 was notably shorter than that of patients with low expression of LINC01436 (Figure 1D). From this, we could conclude that LINC01436 was highly expressed in GC tissues and might take part in promoting cancer progression.
 |
Table 2 Correlation Between LINC01436 and Pathological Parameters in GC |
LINC01436 Regulated the Proliferation, Invasion and Radioresistance of GC Cells
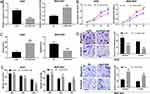
To assess the effect of LINC01436 on GC cells, the LINC01436 low expression and overexpression models were established with AGS cells and BGC-823 cells, respectively (Figure 2A). CCK-8 assay implied that the transfection of LINC01436 siRNA markedly reduced the viability of GC cells, while LINC01436 overexpression notably increased the proliferation of GC cells compared to NC group (Figure 2B). Furthermore, flow cytometry analysis revealed that LINC01436 knockdown enhanced the apoptosis of GC cells, while LINC01436 overexpression exerted the opposite function (Figure 2C). To further elaborate the effect of LINC01436 on GC cell metastasis, Transwell assay was carried out, the results of which unveiled that LINC01436 knockdown reduced GC cell migration and invasion, while LINC01436 overexpression enhanced these processes (Figure 2D). Additionally, colony formation assay implied that LINC01436 knockdown increased the radiosensitivity of GC cells, whereas LINC01436 overexpression had the opposite effect (Figure 2E).
LINC01436 Negatively Modulated miR-513a-5p Expression
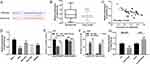
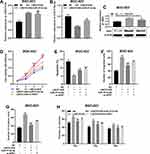
To further probe the downstream mechanism of LINC01436, we turned to StarBase database (http://starbase.sysu.edu.cn/) to predict potential downstream targets of LINC01436 and found that miR-513a-5p was one of its potential targets (Figure 3A). Following that, qRT-PCR was employed to detect the miR-513a-5p expression in GC tissues and corresponding adjacent tissues. The results implied that miR-513a-5p expression was significantly decreased in GC tissues (Figure 3B). Importantly, there was a strong negative correlation between LINC01436 expression and miR-513a-5p expression in the 40 GC tissues mentioned above (Figure 3C). Subsequently, the miR-513a-5p expression in normal cell lines and GC cell lines was detected. As shown, in comparison with the normal cell line GES-1, miR-513a-5p expression was remarkably decreased in GC cell lines (Figure 3D). To further clarify the targeting relationship between LINC01436 and miR-513a-5p, the luciferase activity assay was performed and showed that miR-513a-5p mimics significantly repressed the luciferase activity of wild-type LINC01436 sequence, while overexpression of LINC01436 markedly reversed these effects (Figure 3E). In addition, there was no effect on the mutant LINC01436 sequence (Figure 3E). Consistently, RIP experiments showed that in AGS cells, LINC01436 and miR-513a-5p were significantly enriched in anti-Ago2 group compared with anti-IgG group (Figure 3F). Additionally, qRT-PCR results revealed that LINC01436 overexpression suppressed the miR-513a-5p expression, while LINC01436 knockdown facilitated the expression of miR-513a-5p in GC cells (Figure 3G). Based on these results mentioned above, we identified that miR-513a-5p was a target of LINC01436.
MiR-513a-5p Overexpression Notably Repressed the Proliferation, Invasion and Radioresistance of GC Cells
To delve into the effect of miR-513a-5p on GC cells, the low expression and overexpression models of miR-513a-5p were established (Figure 4A). As shown, miR-513a-5p inhibition enhanced the proliferation of AGS cells, whereas miR-513a-5p mimics restrained it in BGC823 cells (Figure 4B). Flow cytometry analysis indicated that miR-513a-5p inhibition repressed the apoptosis of GC cells, while miR-513a-5p overexpression induced the apoptosis of GC cells (Figure 4C). As we expected, miR-513a-5p inhibitors enhanced the migration and invasion of GC cells, while miR-513a-5p mimics had the opposite effects (Figure 4D). Additionally, colony formation assay revealed that miR-513a-5p inhibitors promoted the radioresistance of GC cells, whereas miR-513a-5p overexpression sensitized GC cell to radiation (Figure 4E). Based on these results, we concluded that miR-513a-5p was a tumor suppressor in GC.
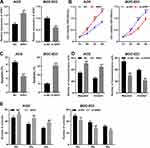
MiR-513a-5p Negatively Regulated APE1 Expression
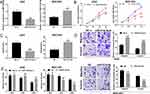
To investigate the APE1 expression in GC, qRT-PCR was employed to detect the APE1 expression in normal tissues and GC tissues. The results uncovered that APE1 expression was markedly up-regulated in GC in comparison with the normal group (Figure 5A). A previous study has confirmed a targeting relationship between miR-513a-5p and APE1.19 Consistent with the previous report, our results found that there was a negative correlation between the expression levels of miR-513a-5p and APE1 in GC tissues (Figure 5B). Western blot was then adopted to detect the APE1 expression after selective regulation of LINC01436 and miR-513a-5p. The results revealed that miR-513a-5p overexpression or LINC01436 knockdown suppressed the APE1 expression, while miR-513a-5p repression or LINC01436 overexpression up-regulated APE1 expression (Figure 5C and D). Therefore, it could be concluded that APE1 expression was negatively regulated by miR-513a-5p, and could be indirectly and positively regulated by LINC01436.
LINC01436/miR-513a-5p/APE1 Axis Modulated the Proliferation, Migration, Invasion and Radioresistance of GC Cells
To figure out the interaction of the LINC01436/miR-513a-5p/APE1 axis in GC, the miR-513a-5p mimics were transfected into BGC-823 cells overexpressing LINC01436. The LINC01436 and miR-513a-5p expressions were detected by qRT-PCR, and the APE1 expression was detected by Western blot. The results revealed that LINC01436 overexpression up-regulated APE1 protein expression and down-regulated miR-513a-5p expression in comparison with the NC group, and miR-513a-5p overexpression down-regulated APE1 expression (vs LINC01436 group) (Figure 6A–C). These results further indicated that LINC01436 could promote the expression of APE1 via repressing miR-513a-5p in GC cells. The results of rescue experiments showed that co-transfection of miR-513a-5p or si-APE1 into BGC-823 cells partly reversed the promotive effects of LINC01436 on the malignant phenotypes of BGC-823 cells (Figure 6D–H). In brief, LINC01436 participated in GC cell proliferation, apoptosis, metastasis and radioresistance via regulating miR-513a-5p and APE1 expressions.
APE1 Silencing Remarkably Suppressed the Proliferation, Invasion and Radioresistance of GC Cells
To explore the biological roles of APE1 on GC cells, the overexpression and knockdown models of APE1 were established with AGS cells and BGC-823 cells, respectively (Figure 7A). Functional assays showed that APE1 overexpression significantly promoted GC cell proliferation but inhibited the cell apoptosis, whereas si-APE1 had the opposite effects in GC cells (Figure 7B and C). Additionally, APE1 overexpression facilitated GC cell migration and invasion, while silencing APE1 exerted opposite effects (Figure 7D). Besides, colony formation assay indicated that APE1 overexpression promoted the radioresistance of GC cells, whereas depleted APE1 sensitized GC cell to radiation (Figure 7E). Collectively, these data suggested that APE1 functioned as a tumor promoter in GC, which was consistent with the previous report.18
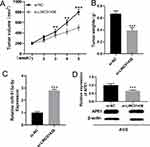
Knockdown of LINC01436 Blocked the Growth of GC Tumors in vivo
In order to further verify the cancer-promoting effect of LINC01436 in GC in vivo, subcutaneous xenograft assay in nude mice was performed. As the result showed, the volume and weight of the tumor in LINC01436 knockdown group were remarkably lower than those in the control group (Figure 8A and B), and the knockdown of LINC01436 up-regulated the expression of miR-513a-5p in the tumor tissues (Figure 8C). Additionally, APE1 expression was detected by qRT-PCR and Western blot in the tumor tissues of mice. The results elucidated that the APE1 expression in tumor tissues of LINC01436 knockdown group was notably lower than that of the control group (Figure 8D). These results further validated the carcinogenic effects of LINC01436 in GC.
Discussion
Currently, standard clinical treatment strategies for GC include surgery, chemotherapy, targeted therapy and radiotherapy. Due to current treatment limitations, most of the GC patients suffer from tumor recurrence, chemoresistance or radioresistance.2 Therefore, in order to find more effective treatments, identifying novel therapy targets of GC is of great significance. Diverse lncRNAs are abnormally expressed in GC and are closely related to the tumorigenesis, proliferation and metastasis of GC, such as lncRNA SNHG6, lncRNA H19, lncRNA DANCR and so on, and they are considered as potential biomarkers and therapy targets.20–22 In this study, we observed that the LINC01436 expression in GC cells was markedly higher than that in the adjacent normal tissues, and the LINC01436 high expression was associated with several pathological features in GC (such as T stage and differentiation status). Importantly, LINC01436 high expression was correlated with poor prognosis of patients. Functional experiments revealed that LINC01436 overexpression remarkably enhanced the malignant phenotypes of GC cells, while knocking down LINC01436 showed the opposite effect. These results fully demonstrated that LINC01436 exerted a cancer-promoting effect in the GC development. To our best knowledge, this was the first study to explore the role of LINC01436 in GC.
In GC, miRNA is endowed with a remarkable regulatory function. For instance, the up-regulation of miR-93 expression facilitates GC cell proliferation, migration, invasion and epithelial-mesenchymal transition to accelerate cancer progression, and miR-532 promotes GC progression via targeting NKD1.23,24 MiR-513a-5p is a miRNA that has been gradually recognized in recent years. Reportedly, miR-513a-5p expression is down-regulated in osteosarcoma tissues and cells, and its overexpression can observably improve the radiosensitivity of osteosarcoma cells.19 In this study, we found that miR-513a-5p expression was down-regulated in GC tissue in comparison with normal gastric tissue. Inhibition of miR-513a-5p enhanced the proliferation, migration and invasion of GC cells, while miR-513a-5p overexpression had the opposite effect, suggesting that miR-513a-5p exerted a tumor-suppressive effect during the GC development.
Accumulating research verifies that lncRNAs act as competitive endogenous RNAs (ceRNAs) to modulate miRNA expression, and the latter regulates protein expression via binding to the 3ʹUTR of mRNAs. This interaction figures prominently in the GC progression.25 For instance, lncRNA MALAT1 acts as a ceRNA to suppress autophagy-mediated chemotherapeutic resistance via inhibiting miR-23b-3p expression;26 LINC01133 modulates APC expression and Wnt/β-catenin pathway via sponging miR-106a-3p to impede GC progression.27 In this work, we found a negative correlation between the LINC01436 expression and miR-513a-5p expression in GC tissues. qRT-PCR, luciferase reporter assay and RIP assay were employed to confirm that LINC01436 could target miR-513a-5p, and LINC01436 overexpression could repress the miR-513a-5p expression in GC cells. In addition, we validated that miR-513a-5p could partially reverse the malignant phenotypes of GC cells promoted by LINC01436 overexpression. Based on these results, we supposed that during GC progression, LINC01436 expression was up-regulated, and then LINC01436, as a kind of ceRNA, suppressed the expression and function of miR-513a-5p, which acted as a tumor suppressor, thereby facilitating the GC progression.
Previous studies reveal that APE1 expression is modulated by microRNA.14,16 For instance, miR-520g and miR-520h can reverse Bortezomib resistance in multiple myeloma via repressing APE1 expression; whereas miR-765 can enhance the anti-angiogenic effect of cisplatin in osteosarcoma via targeting APE1.28,29 In this article, we found that APE1 expression was elevated in GC tissues and its expression was negatively correlated with miR-513a-5p. It is verified that miR-513a-5p can target the regulation of APE1 expression in GC,19 so we are also curious whether APE1 expression can be regulated by LINC01436/miR-513a-5p axis. We found that miR-513a-5p overexpression or LINC01436 knockdown repressed APE1 expression, whereas suppressed miR-513a-5p or LINC01436 overexpression upregulated it. These results reflected that LINC01436 modulated GC progression via modulating the miR-513a-5p/APE1 axis.
Conclusions
In summary, LINC01436 can promote the proliferation, metastasis and radioresistance of GC cells, suggesting that it can be used as a potential therapy target for GC treatment. Mechanism investigation confirms that LINC01436 can regulate GC progression via regulating miR-513a-5p/APE1 axis. Collectively, this study sheds light on the mechanisms of GC progression and provides a new theoretical basis for the diagnosis and treatment of GC.
Author Details
E-mail of co-authors: Ming-dian Lu: [email protected]; Dong Liu: [email protected].
Ethics Approval and Consent to Participate
This study was endorsed by the Ethics Review Committee of First Affiliated Hospital of Anhui Medical University and written informed consent was obtained from each patient.
Author Contributions
All authors made great contributions to the experimental concept and design, data acquisition or data analysis and interpretation; participated in drafting the article or strictly revised the content of important knowledge content; agreed on the journal to which the article will be submitted; gave final approval of the version to be published. All authors have agreed to be responsible for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
References
1. Choi YJ, Kim N. Gastric cancer and family history. Korean J Intern Med. 2016;31(6):1042–1053. doi:10.3904/kjim.2016.147
2. Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626. doi:10.1177/1010428317714626
3. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
4. Li PF, Chen SC, Xia T, et al. Non-coding RNAs and gastric cancer. World J Gastroenterol. 2014;20(18):5411–5419. doi:10.3748/wjg.v20.i18.5411
5. Huang HW, Xie H, Ma X, Zhao F, Gao Y. Upregulation of LncRNA PANDAR predicts poor prognosis and promotes cell proliferation in cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21(20):4529–4535.
6. Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51. doi:10.1101/gad.270959.115
7. Yuan S, Xiang Y, Wang G, et al. Hypoxia-sensitive LINC01436 is regulated by E2F6 and acts as an oncogene by targeting miR-30a-3p in non-small cell lung cancer. Mol Oncol. 2019;13(4):840–856. doi:10.1002/1878-0261.12437
8. Alessandrini L, Manchi M, De Re V, Dolcetti R, Canzonieri V. Proposed molecular and miRNA classification of gastric cancer. Int J Mol Sci. 2018;19(6):1683. doi:10.3390/ijms19061683
9. Yang W, Ma J, Zhou W, et al. Molecular mechanisms and theranostic potential of miRNAs in drug resistance of gastric cancer. Expert Opin Ther Targets. 2017;21(11):1063–1075. doi:10.1080/14728222.2017.1389900
10. Fan Z, Cui H, Xu X, et al. MiR-125a suppresses tumor growth, invasion and metastasis in cervical cancer by targeting STAT3. Oncotarget. 2015;6(28):25266–25280. doi:10.18632/oncotarget.4457
11. Shi L, Wang Y, Lu Z, et al. miR-127 promotes EMT and stem-like traits in lung cancer through a feed-forward regulatory loop. Oncogene. 2017;36(12):1631–1643. doi:10.1038/onc.2016.332
12. Shin S, Moon KC, Park KU, Ha E. MicroRNA-513a-5p mediates TNF-α and LPS induced apoptosis via downregulation of X-linked inhibitor of apoptotic protein in endothelial cells. Biochimie. 2012;94(6):1431–1436. doi:10.1016/j.biochi.2012.03.023
13. Muti P, Donzelli S, Sacconi A, et al. MiRNA-513a-5p inhibits progesterone receptor expression and constitutes a risk factor for breast cancer: the hOrmone and Diet in the ETiology of breast cancer prospective study. Carcinogenesis. 2018;39(2):98–108. doi:10.1093/carcin/bgx126
14. Shah F, Logsdon D, Messmann RA, et al. Exploiting the Ref-1-APE1 node in cancer signaling and other diseases: from bench to clinic. NPJ Precis Oncol. 2017;1.
15. Whitaker AM, Freudenthal BD. APE1: a skilled nucleic acid surgeon. DNA Repair (Amst). 2018;71:93–100. doi:10.1016/j.dnarep.2018.08.012
16. Antoniali G, Serra F, Lirussi L, et al. Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism. Nat Commun. 2017;8(1):797. doi:10.1038/s41467-017-00842-8
17. Chen T, Liu C, Lu H, et al. The expression of APE1 in triple-negative breast cancer and its effect on drug sensitivity of olaparib. Tumour Biol. 2017;39(10):1010428317713390. doi:10.1177/1010428317713390
18. Qing Y, Li Q, Ren T, et al. Upregulation of PD-L1 and APE1 is associated with tumorigenesis and poor prognosis of gastric cancer. Drug Des Devel Ther. 2015;9:901–909. doi:10.2147/DDDT.S75152
19. Dai N, Qing Y, Cun Y, et al. miR-513a-5p regulates radiosensitivity of osteosarcoma by targeting human apurinic/apyrimidinic endonuclease. Oncotarget. 2018;9(39):25414–25426. doi:10.18632/oncotarget.11003
20. Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42(3):999–1012. doi:10.1159/000478682
21. Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi:10.18632/oncotarget.1913
22. Mao Z, Li H, Du B, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37(6). doi:10.1042/BSR20171070.
23. Guan H, Li W, Li Y, et al. MicroRNA-93 promotes proliferation and metastasis of gastric cancer via targeting TIMP2. PLoS One. 2017;12(12):e0189490. doi:10.1371/journal.pone.0189490
24. Hu S, Zheng Q, Wu H, et al. miR-532 promoted gastric cancer migration and invasion by targeting NKD1. Life Sci. 2017;177:15–19. doi:10.1016/j.lfs.2017.03.019
25. Mao Y, Liu R, Zhou H, et al. Transcriptome analysis of miRNA-lncRNA-mRNA interactions in the malignant transformation process of gastric cancer initiation. Cancer Gene Ther. 2017;24(6):267–275. doi:10.1038/cgt.2017.14
26. YiRen H, YingCong Y, Sunwu Y, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16(1):174. doi:10.1186/s12943-017-0743-3
27. Yang XZ, Cheng TT, He QJ, et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17(1):126.
28. Yuan X, Ma R, Yang S, et al. miR-520g and miR-520h overcome bortezomib resistance in multiple myeloma via suppressing APE1. Cell Cycle. 2019;18(14):1660–1669. doi:10.1080/15384101.2019.1632138
29. Liang W, Wei X, Li Q, et al. MicroRNA-765 enhances the anti-angiogenic effect of CDDP via APE1 in osteosarcoma. J Cancer. 2017;8(9):1542–1551. doi:10.7150/jca.18680
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.