Back to Journals » OncoTargets and Therapy » Volume 14
LINC01278 is Highly Expressed in Osteosarcoma and Participates in the Development of Tumors by Mediating the miR-134-5p/KRAS Axis
Authors Zhang GF, Zhou BS, An XC, An FM, Li SH
Received 1 June 2020
Accepted for publication 8 December 2020
Published 26 January 2021 Volume 2021:14 Pages 683—695
DOI https://doi.org/10.2147/OTT.S265591
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Guo-Feng Zhang, Bai-Sui Zhou, Xiao-Chun An, Feng-Min An, Shan-Hui Li
Department of Orthopedics, Yantai Affiliated Hospital of Binzhou Medical University, Yantai 261400, People’s Republic of China
Correspondence: Guo-Feng Zhang
Department of Orthopedics, Yantai Affiliated Hospital of Binzhou Medical University, No. 717, Jinbu Street, Yantai 264100, Shandong, People’s Republic of China
Tel +86-15905351140
Fax +86 535-4770841
Email [email protected]
Purpose: There is increasing evidence that non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), produce a critical regulatory effect on osteosarcoma (OS). LINC01278, as a newly discovered lncRNA, is found to be highly expressed in OS, but its related mechanism remains unclear. This research, therefore, is designed to study the mechanism of LINC01278 in OS and to find potential targets for clinical use.
Methods: qRT-PCR was applied to determine the relative expression of LINC01278 and analyze its diagnostic value in OS. CCK-8, Transwell and flow cytometry were utilized for the determination of cell proliferation, migration/invasion, and apoptosis. RIP and RNA pull-down experiments were used to verify the targeted binding effect of miR-134-5p and LINC01278. The relationship between miR-134-5p and LINC01278 or KRAS was analyzed using dual luciferase reporter gene. The effects of LINC01278 on tumor growth in nude mice was analyzed by in vivo experiment.
Results: qRT-PCR showed that LINC01278 increased in OS tissues and serum, indicating poor prognosis. In addition, LINC01278 was also of high value for OS diagnosis. Functional experiments showed that LINC01278 inhibited KRAS-mediated OS cell proliferation and metastasis through miR-134-5p. Finally, the results of an in vivo animal model indicated that LINC01278 promoted OS growth.
Conclusion: LINC01278 is expressed highly in OS, and patients with high LINC01278 expression have poor prognosis. Moreover, LINC01278 can suppress the proliferation and apoptosis of OS cells through mediating miR-134-5p/KRAS axis, which is expected to become a potential therapeutic target for OS.
Keywords: LINC01278, miR-134-5p, KRAS, osteosarcoma, proliferation, apoptosis
Introduction
Osteosarcoma (OS) tends to occur in mesenchymal tissues and accounts for 0.2% of all human malignant tumors, most often in adolescents aged 10–20 years.1 Research shows that 70% of OS patients are adolescents.2 OS mainly occurs in the metaphyseal region with more active production and is prone to lung metastasis in the early stage due to its extremely rapid growth.3 At present, the main clinical treatment plan for OS is the combination of surgery and chemotherapy. However, due to the characteristics of easy metastasis of diseases, patients may have metastasis during the treatment process, which will inevitably increase the difficulty of treatment and affect the prognosis and survival of patients.4,5 Hence, it is of great importance to study the pathogenesis of OS, and deeply understand the progression and development of diseases and target therapy.
For the past few years, the research on long non-coding RNA (LncRNA) has been deepened. LncRNA is a variety of non-coding single-stranded RNA with a length of more than 200nt.6 At first, scholars believed that lncRNA has no ability to encode proteins. However, with the discovery of various functions of lncRNA in recent years, it is found that lncRNA has epigenetic changes, transcriptional regulation and post-transcriptional modification. In addition, it is involved in the progression and development of various diseases through targeted regulation of microRNA and target proteins.7,8 Previous research has confirmed that lncRNA produces an important regulatory effect on the progression of OS.9 For example, in the study of Chen et al10 lncRNA TP73-AS1 can predict the adverse prognosis of OS and cause OS. In the research of Guo et al11 lncRNA-SRA1 can inhibit OS cell proliferation and promote cell apoptosis. LINC01278, as a member of lncRNA family, is a newly discovered lncRNA. LINC01278 has been found to be highly expressed in breast cancer and can inhibit the development of papillary thyroid carcinoma by regulating microRNA (miR)-376c-3p/DNM3 axis.12 However, there are relatively few researches on OS. Previous research found that LINC01278 can participate in OS development by regulating the miR-134-5p/PTH1 axis. According to this research, it was found that LINC01278 and miR-134-5p have targeted relationship through multiple online prediction websites. There is a study saying that miR-134-5p is poorly expressed in OS and can participate in OS development by mediating downstream target genes.13
Therefore, we aimed to explore the relevant mechanisms of LINC01278 and miR-134-5p in OS, and provide potential targets for clinical treatment in this study.
Methods and Data
Tissue Samples
A total of 66 cases of OS patients treated in Yantai Affiliated Hospital of Binzhou Medical University from January 2010 to January 2014 were collected. Cancer tissues and paracancerous tissues of the patients were collected during the operation, transported with liquid nitrogen and sent to the laboratory for testing. The tissues were stored at −80°C. Peripheral blood (3 mL) was collected from patients preoperatively, and peripheral blood (3 mL) was also collected from 66 cases of healthy subjects simultaneously. The obtained peripheral blood was placed in the test tube. The samples were incubated at 37°C for 1 hour and centrifuged at 1500 x g at 4°C for 15 minutes immediately for serum collection and analysis of the diagnostic value of LINC01278 in OS. All patients enrolled were those who had received treatment for the first time and were confirmed as OS by preoperative biopsy and pathological examination (via postoperative collection of tumor tissues), and each patient signed an informed consent form. With the approval of the Medical Ethics Committee of Yantai Affiliated Hospital of Binzhou Medical University, this study was performed in strict accordance with the Declaration of Helsinki.14
Cell Culture
OS cells SAOS-2, MG63, U2OS, SJSA1 and human osteoblast hFOB 1.19 were bought from the American Type Culture Collection (Manassas, VA, USA). RPMI-1640 medium containing 10% fetal bovine serum (FBS), 50 U/mL penicillin and 50 μg/mL streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) was applied for cell culture under 37°C and 5% CO2 in a constant temperature incubator.
Cell Transfection
Design and synthesis of the short hairpin RNA (shRNA) vector or over-expression vector of LINC01278/KRAS and miR-134-5p mimics/inhibitor were carried out by GenePharma in Shanghai, People's Republic of China. Six μg/mL polyvinylamine (SIGMA) and 1×106 recombinant lentivirus transduction units infected 60~80% of the fused cells. After 48 hours of stable transfection, the collected cells were treated with 2 μg/mL puromycin for 2 weeks and detected by flow cytometry (CytoFLEX LX, Beckman Kurt, USA). In this study, cell transfection was performed using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, USA) ki. The transfection procedure was performed according to the kit instructions.
Quantitative Reverse‐Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from the collected samples (cells, tissues, serum) with TRIzol reagent (Invitrogen, USA). UV spectrophotometer (NanoDrop ND-2000, Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis were utilized to detect the purity, concentration and integrity of the extracted total RNA. Total RNA (1 μg) was reverse transcribed into cDNA with a final volume of 20 μL using PrimeScript RT kit (Takara, Dalian, People's Republic of China) under standard conditions with random primers. The SYBR PreMix Ex Taq (Takara, Beijing, People's Republic of China) kit and ABI 7500PCR instrument (ABI, USA) were used for PCR detection. The operation procedures were performed according to the kit instructions. U6 was taken as the internal reference of miR, and GAPDH was taken as the internal reference of lncRNA and mRNA. Data collection was based on the comparison period threshold (CT) method (2−ΔΔCT).15 Primer sequence: LINC01278 upstream primer: 5ʹ-AGGTTCCCCAGTCCCTATGTG-3ʹ, downstream primer: 5ʹ-TCTACAGGGAGTGCAAGGGT-3ʹ. miR-134-5p upstream primer: 5ʹ-GAAGCTCATTGGAGACCCTAAC-3ʹ, downstream primer: 5ʹ-CAACCTCTAAGTCGTGCTCATAC-3ʹ. KRAS upstream primer: 5ʹ-TCTCCTTCTCAGGATTCCTACAG-3ʹ, downstream primer: 5ʹ-ACAAAGAAAGCCCTCCCCAGT-3ʹ. GAPDH upstream primer: 5ʹ-TAATCTTCGCCTTAATACTT-3ʹ, downstream primer: 5ʹ-AGCCTTCATACATCTCAA-3ʹ. U6 upstream primer: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, downstream primer: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Cell Proliferation Detection
The cell counting kit-8 (CCK-8, Dojindo, Kumamoto, Japan) was applied to evaluate the cell viability. The method was as follows: the transfected cells were collected, resuspended, adjusted to 3×103 cells/well, and transferred to 96-well plates. Each well was inoculated with 100 μL of α-MEM medium containing 10% FBS. CCK-8 solution was put into each well at 0, 24, 48, and 72 hours, respectively, and then the cells were cultured at 37°C for 1.5 hours. Subsequently, the absorbance was measured at 450 nm. Cell proliferation was detected in the light of the instructions of the kit.
Cell Apoptosis and Cell Cycle Analysis
The transfected cells were collected, digested with trypsin, rinsed twice with phosphate buffer saline (PBS), added with 500 μL binding buffer, resuspended and transferred to flow tube. Then, they were added with AnnexinV-FITC (5 μL) and PI (5 μL) in sequence, incubated in the dark at indoor temperature for 5 minutes, and detected with Beckman Kurt CytoFLEX LX flow cytometer system. The experiment was repeated 3 times and averaged. Transfected cells were collected and rinsed twice with PBS. PBS (200 μL) was added to resuspend the cells to form cell suspension, and 70% ethanol was added to fix the cells at 4°C for 2 hours. Centrifuge was performed at 1000 r/min for 5 min. Then the cells were rinsed twice with PBS, added with PI dye liquor, and incubated in the dark for 30 minutes. The changes of cell cycle in each experimental group were measured using flow cytometry and Single Histogram Statistic analysis software.
WB Assay
Cells in each group were collected after transfection, and the total protein was extracted by RIPA lysis (Beyotime Institute of Biotechnology, Haimen, People's Republic of China). The protein concentration was detected by BCA assay (Pierce, Thermo Fisher Scientific, Inc.), adjusted to 4 μg/μL, separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membrane. Then, it was stained with Ponceau S and washed after soaking in phosphate buffered solution (PBST) for 5 min. It was sealed with 5% skimmed milk powder for 2 hours, and then primary antibody (1: 1000) was added to it to seal at 4°C for one night. The membrane was washed to remove primary antibody, and HRP-labeled goat anti-rabbit IgG (1:500) secondary antibody (1:5000) was put into it for incubation at 37°C for 1 hour. It was rinsed with PBS for 3 times, with 5 minutes each time. Excess liquid was absorbed from the membrane with filter paper. Electrochemiluminescence (ECL) was used to illuminate and develop in a dark room. Protein bands were scanned, and the grayscale value was analyzed in Quantity One software. Relative expression level of the protein = grayscale value of the target protein band/β-actin protein band. Both primary and secondary antibodies were from Abcam (Cambridge, UK).
Dual Luciferase Reporter
DNA oligonucleotide and pMiR-Reporter Vector were utilized to construct report vector of LINC01278 wild type/mutant (LINC01278-WT/MUT) and KRAS wild type/mutant (KRAS-WT/MUT). Subsequently, the above two vectors were co-transfected with miR-150-5p-mimics and negative control (miR-NC), respectively, and then co-transfected into HEK293 cells. After incubation for 24 hours, the collected cells were tested for luciferase activity by the aid of a dual luciferase reporter kit (Promega, USA).
RNA Immunoprecipitation (PIR)
RIP experiments were conducted by EZ-Magna RIP kit (Millipore, USA), and the experimental procedures were strictly in accordance with the kit instructions. The specific steps of RCC cell collection were as follows: cells were lysed using RIPA, and then the whole cell protein extract was incubated with RIP washing buffer comprising magnetic beads bound to anti-AgO2 antibody (Millipool) or mouse immunoglobulin G (IgG) control. Protease K was used to digest the sample protein, the immunoprecipitated RNA was extracted, and analysis of purified RNA was performed by qRT-PCR to prove the existence of binding target.
RNA Pull-Down
EC109 cells were transfected with biotinylated miR-134-5p-wt, miR-134-5p-mut and negative control (GenePharma, Shanghai, People's Republic of China), respectively. Forty-eight hours later, the cell lysate was incubated with M-280 streptomycete magnetic beads (Invitrogen) in the light of the production instructions. Then qRT-PCR was used to estimate LINC01278 level in the RNA complex bound to the beads.
In vivo Experiments
Fifteen BALB/c-nu nude mice (5–6 weeks old, 18–20 g, Beijing Vital River Laboratory Animal Technology Co., Ltd., People's Republic of China) were purchased and randomly divided into three groups, with five mice in each group. They were inoculated with U2OS cells with high expressed LINC01278 and suppressed expressed U2OS (1×107 cells in 100 μL serum-free medium), respectively, and the other group was the control group. Tumor size was measured with calipers every 7 days after NC transfection, and the formula was (length× width2)/2 (mm3). Animals were euthanized 28 days later, and tumors were removed and weighed. This study was approved by the Animal Ethics Committee of Yantai Affiliated Hospital of Binzhou Medical University and was conducted in the light of the Guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health.
Statistical Analysis
SPSS v. 20.0 software package (IBM Corp., Armonk, NY, USA) was applied for statistical analysis of the collected data, and GraphPad 7 software package (SanDiego, CA, USA) was used to draw the required images. The independent sample t-test was used for intergroup comparisons. Comparison among groups were analyzed via one-way analysis of variance, represented with F. Least-Significant Difference t-test was used for afterwards pairwise comparison. Comparison among multiple time points were analyzed using Repeated Measures, denoted by F. Bonferroni was used for back testing. ROC was used to draw the curve of diagnostic value of lncRNA in OS. Kaplan-Meier survival curve was utilized when plotting the 5-year survival of patients, which was analyzed by Log rank test. Prognosis of patients was analyzed using multivariate Cox regression analysis. When P<0.05, there was a statistical difference.
Results
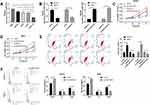
LINC01278 is Highly Expressed in OS and is a Prognostic Indicator of OS
The expression of LINC01278 in patient’s tissues and serum was detected separately, which was remarkably higher in patients’ tissues than in paracancerous tissues (Figure 1A). And its expression in serum was also observed to increase in patients’ serum (Figure 1B). Pearson test revealed that LINC01278 expression in serum was positively correlated with the expression in tissues (Figure 1C). According to the median value of LINC01278 expression, patients were divided into a high expression group and a low expression group. By observing the association between LINC01278 and pathological data of OS patients, we found that tumor size, clinical stage and distal metastasis of patients were correlated with LINC01278, while gender, age and tumor location of patients were not correlated with LINC01278 (Table 1). Further follow-up showed that the patients with low expression of LINC01278 had higher 5-year survival rates (Figure 1D). In addition, Cox regression analysis showed that LINC01278 was an independent prognostic factor for OS patients (Table 2).
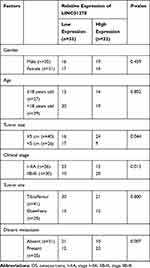 |
Table 1 Relationship Between LINC01278 and Clinical Data of OS Patients |
 |
Table 2 Cox Regression Analysis |
The Value of LINC01278 in Diagnosing OS
In order to further determine the diagnostic value of LINC01278 in OS, the expression of LINC01278 in serum of OS patients was analyzed. The results showed that the area under the curve (AUC) of LINC01278 in distinguishing OS patients from normal persons was 0.945 (Figure 2A). Then we further observed the diagnostic value of LINC01278 in tumor size, clinical stage and distal metastasis. The results showed that the AUC of LINC01278 in distinguishing tumors ≥ 3cm and < 3 cm was 0.806 (Figure 2B), the AUC of it in distinguishing Phase I–IIA and phase IIB-III was 0.7361 (Figure 2C), and the AUC of it in distinguishing distal metastasis was 0.6737 (Figure 2D), which suggested that LINC01278 might be a potential indicator for diagnosing OS (Table 3).
 |
Table 3 ROC Parameters of LINC01278 in Diagnosing Clinical Indicators |
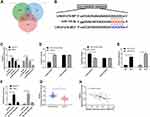
Knock-Down LINC01278 Inhibits OS Cell Proliferation and Apoptosis
In order to determine the impacts of regulating LINC01278 on OS cells, high expression of LINC01278 was first determined in OS cells (Figure 3A). Then, cell transfection was performed on vectors whose LINC01278 were up-regulated and knocked down (Figure 3B). Through observation, CCK-8 experiment exhibited that the proliferation ability of cells transfected with sh-LINC01278 was gradually inhibited with the increase of time compared with those of transfected with sh-NC (Figure 3C and D). Flow cytometry indicated that the apoptosis rate of sh-LINC01278 transfected cells was notably higher than that of sh-NC transfected cells (Figure 3E). The cell cycle analysis showed that the number of cells in the G1/G0 phase increased significantly after transfection with sh-LINC01278, while in the S and G2/M phases it decreased, and the results were reversed after up-regulation of LINC01278 (Figure 3F). This suggested that knocking down LINC01278 could induce cell apoptosis, arrest cell cycle at G1/G0, and further inhibit cell proliferation.
LINC01278 Can Act as a Sponge for miR-134-5p
In this study, in order to determine whether LINC01278 could specifically bind miR, we used online prediction websites starBase,16 miRDB,17 and LncBase18 to make predictions. Through predictions, a total of 19 potentially collectible miRs were found (Figure 4A and B). Combined with existing literature, miR-134-5p was elected for analysis. First, miR-134-5p expression in cells transfected with pcDNA-LINC01278 and sh-LINC01278 was found to be significantly inhibited and increased (Figure 4C). Further dual luciferase reporter confirmed that miR-134-5p-mimics could inhibit the fluorescence activity of LINC01278-WT (Figure 4D). RIP experiments also revealed that miR-134-5p and LINC01278 could be precipitated by Ago2 protein (Figure 4E). RNA pull-down experiment confirmed that LINC01278 increased only in the complex that was lowered by Bio-miR-134-5p-WT, rather than in Bio-NC and Bio-miR-134-5p-Mut (Figure 4F). Finally, miR-134-5p expression was also found to be low in OS patient tissues (Figure 4G). Correlation analysis suggested that there was a negative correlation between miR-134-5p and LINC01278 (Figure 4H), indicating that LINC01278 might specifically bind miR-134-5p to participate in OS progress.
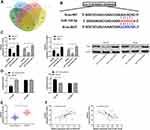
miR-134-5p Can Target KRAS
miR participating in the occurrence of tumors by regulating downstream target genes has been confirmed by many studies. In order to identify the downstream target genes of miR-134-5p, we predicted them through starBase, miRBD, TargetScan,19 and miRTarBase20 online websites (Figure 5A and B). As a result, a total of three potential target binding genes (KRAS, SMAD6, STAT5B) were found, and KRAS was selected for research in combination with existing literature. By detecting the cells after transfection of miR-134-5p-mimics and miR-134-5p-inhibit, the relative expression of KRAS mRNA and protein was found to be notably inhibited and improved (Figure 5C), and the dual luciferase reporter confirmed that miR-134-5p-mimics could effectively inhibit KRAS-WT fluorescence activity (Figure 5D). By detecting KRAS expression in OS patient tissues, it could be seen that the relative expression of KRAS in tumor tissues of patients increased significantly (Figure 5E). And the correlation analysis found that miR-134-5p was negatively correlated with KRAS, while KRAS was positively correlated with LINC01278 expression (Figure 5F). This indicated that LINC01278 might participate in OS growth through acting as a sponge of miR-134-5p to regulate KRAS expression.
LINC01278 Inhibits Proliferation and Apoptosis of OS Cells by Mediating miR-134-5p/KRAS Axis
In order to confirm whether LINC01278 affected the proliferation and apoptosis of OS cells through the miR-134-5p/KRAS axis, we co-transfected some vectors. Through our experiments, it was found that the proliferation ability of cells transfected with miR-134-5p-inhibit and pcDNA-KRAS was remarkably improved (Figure 6A), and the apoptosis was remarkably inhibited (Figure 6B). The number of cells in G1/G0 phase was remarkably decreased (Figure 6C), but was increased in S and G2/M phase. However, when sh-LINC01278 was co-transfected with miR-134-5p-inhibit and pcDNA-KRAS separately, the cell proliferation ability, apoptosis rate and cell cycle number were reversed, without significant difference when compared with sh-NC. This indicated that LINC01278 could suppress the proliferation and apoptosis of OS cells through mediating the miR-134-5p/KRAS axis.
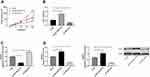
Effects of LINC01278 Regulation on OS Growth
At the end of the study, we conducted an in vivo experiment. Through the experiment, it was found that the tumor volume and mass of nude mice injected with pcDNA-LINC01278 were remarkably elevated compared with nude mice injected with sh-NC (Figure 7A), while the tumor volume and mass of nude mice injected with sh-LINC01278 were significantly decreased (Figure 7B). In addition, we detected miR-134-5p and KRAS expression in nude mice tumors, and discovered that miR-134-5p expression was inhibited and KRAS was significantly elevated in nude mice tumor tissues after injection of pcDNA-LINC01278 (Figure 7C and D), while the expression of miR-134-5p was increased and KRAS was inhibited in nude mice tumor tissues after injection of sh-LINC01278.
Discussion
OS, as a common orthopedic malignant tumor in clinic, most often occurring in adolescents, has a high incidence and is prone to metastasis.21 In this paper, LINC01278 was found to be expressed highly in OS patients, and the prognosis of patients with high expression was poor. In addition, LINC01278 had certain clinical value in the diagnosis of OS and its clinically related indicators, suggesting that LINC01278 was expected to become a potential observation indicator of OS.
LINC01278, as a newly discovered lncRNA, is located in human Xq11.1. There have been very few previous studies on LINC01278. In this study, we confirmed that LINC01278 is highly expressed in OS patients through clinical research, and the prognosis of patients with high expression is poor. In addition, LINC 01278 has high value in the diagnosis of OS. In order to further observe the effect of LINC01278 on OS, we conducted in vitro experiments. The results showed that by knocking down LINC01278, G0/G1 phase was blocked, cell proliferation and apoptosis were inhibited, and the results were reversed after over-expression of LINC01278, suggesting that LINC01278 is a promising target in treating OS.
ceRNA was first proposed by Leonardo Salmena et al,22 that is, ceRNA is an endogenous sponge that can affect the distribution of miRNAs on its target, thus exerting another layer of new post-transcriptional regulation. Previous studies have found that lncRNA can be used as ceRNA of miR. For example, studies by Cui et al23 have found that lncRNA UCA1 regulates the progression of colon cancer by regulating the miR-28-5p/HOXB3 axis. Other studies have found that lncRNA PSMG3AS1 acts as an miR1433p sponge to increase the proliferation and migration of breast carcinoma cells.24 In order to explore the mechanism of LINC01278, an online prediction website predicted that miR-134-5p and LINC01278 do have specific binding sites. As a member of the miR family, miR-134-5p is shown to to have low expression in lung cancer,25 gastric cancer,26 retinoblastoma,27 and other tumors. Moreover, some studies have shown that miR-134-5p inhibits OS cell invasion and metastasis by regulating MMP1 and MMP3.28 Therefore, in this research, miR-134-5p expression in OS patients was detected firstly. The results indicated that miR-134-5p has low expression low in OS patients, and correlation analysis demonstrated a negative correlation between miR-134-5p and LINC01278, suggesting that LINC01278 might target miR-134-5p. To verify the conjecture, we carried out verification tests. Through a dual luciferase reporter, RIP and RNA pull-down experiment, it was confirmed that LINC01278 could specifically bind to miR-134-5p and regulate its expression in a target manner.
miR has been proven to inhibit tumor growth and metastasis by targeting downstream target genes.29 Prediction of miR-134-5p downstream target genes through online prediction websites exhibited that KRAS and miR-134-5p have potential targeted binding relationships. KRAS is Kirsten ras oncogene homologous gene of mammalian ras gene family.30 Previous studies have shown that abnormal activation of KRAS protein changes normal RAS/PI3K/AKT signals and influences cell proliferation, invasion and apoptosis.31,32 In this paper, we detected the KRAS expression in OS patient tissues and found that it is expressed highly in OS tissues, showing a negative correlation with miR-134-5p and a positive correlation with LINC01278, which suggests that LINC01278 may regulate KRAS expression and participate in OS growth by acting as a sponge of miR-134-5p. The targeted relationship between miR-134-5p and KRAS was confirmed through dual luciferase reporter. Then, the proliferation and apoptosis of cells transfected with miR-134-5p-inhibit and pcDNA-KRAS were observed. After transfection, cell proliferation and apoptosis were significantly inhibited, but when we co-transfected sh-LINC01278 with miR-134-5p-inhibit and pcDNA-KRAS, separately, the proliferation and apoptosis were reversed. This showed that LINC01278 could suppress the proliferation and apoptosis of OS cells by mediating the miR-134-5p/KRAS axis. In addition, we also found through in vivo experiments that the tumor volume and mass of nude mice injected with pcDNA-LINC01278 significantly increased, miR-134-5p expression in tumor tissue was inhibited, and KRAS expression increased, while the results of nude mice injected with sh-LINC01278 were reversed. This further supports our conclusion.
Conclusion
LINC01278 is expressed highly in OS, and the prognosis of patients with high expression is poor. Moreover, LINC01278 can suppress the proliferation and apoptosis of OS cells through mediating the miR-134-5p/KRAS axis, which is expected to become a potential therapeutic target for OS (Figure 8).
 |
Figure 8 Schematic diagram of LINC01278/miR-134-5p/KRAS in OS. |
Disclosure
The authors report no conflicts of interest in this work.
References
1. Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92.
2. Levy M, Leclerc BS. Fluoride in drinking water and osteosarcoma incidence rates in the continental United States among children and adolescents. Cancer Epidemiol. 2012;36(2):e83–e88. doi:10.1016/j.canep.2011.11.008
3. Liu Q, Yang G, Qian Y. Loss of MicroRNA-489-3p promotes osteosarcoma metastasis by activating PAX3-MET pathway. Mol Carcinog. 2017;56(4):1312–1321. doi:10.1002/mc.22593
4. Wang Y, Guo W, Shen D, et al. Surgical treatment of primary osteosarcoma of the sacrum: a case series of 26 patients. Spine (Phila Pa 1976). 2017;42(16):1207–1213. doi:10.1097/BRS.0000000000002043
5. Wu MS, Ma QY, Liu DD, et al. CDC20 and its downstream genes: potential prognosis factors of osteosarcoma. Int J Clin Oncol. 2019;24(11):1479–1489. doi:10.1007/s10147-019-01500-3
6. Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–116. doi:10.1093/bib/bbv031
7. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi:10.1016/j.bbagrm.2014.08.012
8. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–286.
9. Zhu C, Cheng D, Qiu X, Zhuang M, Liu Z. Long noncoding RNA SNHG16 promotes cell proliferation by sponging microRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell Physiol Biochem. 2018;51(1):429–440. doi:10.1159/000495239
10. Chen X, Zhou Y, Liu S, et al. LncRNA TP73-AS1 predicts poor prognosis and functions as oncogenic lncRNA in osteosarcoma. J Cell Biochem. 2018.
11. Guo W, Jiang H, Li H, et al. LncRNA-SRA1 suppresses osteosarcoma cell proliferation while promoting cell apoptosis. Technol Cancer Res Treat. 2019;18:1533033819841438. doi:10.1177/1533033819841438
12. Lin S, Tan L, Luo D, Peng X, Zhu Y, Li H. Linc01278 inhibits the development of papillary thyroid carcinoma by regulating miR-376c-3p/DNM3 axis. Cancer Manag Res. 2019;11:8557–8569. doi:10.2147/CMAR.S217886
13. Fu D, Lu C, Qu X, et al. LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY). 2019;11(19):8374–8385. doi:10.18632/aging.102325
14. General Assembly of the World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. J Am Coll Dent. 2014;81(3):14–18.
15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
16. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D97. doi:10.1093/nar/gkt1248
17. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi:10.1093/nar/gkz757
18. Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi:10.1093/nar/gkv1270
19. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. doi:10.7554/eLife.05005
20. Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi:10.1093/nar/gkx1067
21. Shimose S, Kubo T, Fujimori J, Furuta T, Ochi M. A novel assessment method of serum alkaline phosphatase for the diagnosis of osteosarcoma in children and adolescents. J Orthop Sci. 2014;19(6):997–1003. doi:10.1007/s00776-014-0617-0
22. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
23. Cui M, Chen M, Shen Z, Wang R, Fang X, Song B. LncRNA-UCA1 modulates progression of colon cancer through regulating the miR-28-5p/HOXB3 axis. J Cell Biochem. 2019;120:6926–6936. doi:10.1002/jcb.27630
24. Cui Y, Fan Y, Zhao G, et al. Novel lncRNA PSMG3AS1 functions as a miR1433p sponge to increase the proliferation and migration of breast cancer cells. Oncol Rep. 2020;43(1):229–239. doi:10.3892/or.2019.7390
25. Zhang L, Huang P, Li Q, Wang D, Xu CX. miR-134-5p promotes stage I lung adenocarcinoma metastasis and chemoresistance by targeting DAB2. Mol Ther Nucleic Acids. 2019;18:627–637. doi:10.1016/j.omtn.2019.09.025
26. Chi J, Liu T, Shi C, et al. Long non-coding RNA LUCAT1 promotes proliferation and invasion in gastric cancer by regulating miR-134-5p/YWHAZ axis. Biomed Pharmacother. 2019;118:109201. doi:10.1016/j.biopha.2019.109201
27. Wang H, Ji X. SMAD6, positively regulated by the DNM3OS-miR-134-5p axis, confers promoting effects to cell proliferation, migration and EMT process in retinoblastoma. Cancer Cell Int. 2020;20:23. doi:10.1186/s12935-020-1103-8
28. Chen CL, Zhang L, Jiao YR, et al. miR-134 inhibits osteosarcoma cell invasion and metastasis through targeting MMP1 and MMP3 in vitro and in vivo. FEBS Lett. 2019;593(10):1089–1101. doi:10.1002/1873-3468.13387
29. McManus DD, Rong J, Huan T, et al. Messenger RNA and MicroRNA transcriptomic signatures of cardiometabolic risk factors. BMC Genom. 2017;18(1):139. doi:10.1186/s12864-017-3533-9
30. Lampson BL, Pershing NL, Prinz JA, et al. Rare codons regulate KRas oncogenesis. Curr Biol. 2013;23(1):70–75. doi:10.1016/j.cub.2012.11.031
31. Vitiello PP, Cardone C, Martini G, et al. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J Exp Clin Cancer Res. 2019;38(1):41. doi:10.1186/s13046-019-1035-0
32. Takino JI, Sato T, Nagamine K, Hori T. The inhibition of Bax activation-induced apoptosis by RasGRP2 via R-Ras-PI3K-Akt signaling pathway in the endothelial cells. Sci Rep. 2019;9(1):16717. doi:10.1038/s41598-019-53419-4
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.