Back to Journals » OncoTargets and Therapy » Volume 13
LINC01272 Promotes Migration and Invasion of Gastric Cancer Cells via EMT
Authors Leng X , Liu G, Wang S, Song J , Zhang W , Zhang X, Rong L, Ma Y, Song F
Received 12 December 2019
Accepted for publication 27 March 2020
Published 22 April 2020 Volume 2020:13 Pages 3401—3410
DOI https://doi.org/10.2147/OTT.S242073
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Xue Leng,1 Geli Liu,1 Sen Wang,2 Jing Song,3 Wanfeng Zhang,3 Xianqin Zhang,1 Li Rong,1,4 Yongping Ma,1 Fangzhou Song1
1Molecular Medicine and Cancer Research Center, Chongqing Medical University, Chongqing 400016, People’s Republic of China; 2Laboratory Department, Affiliated Hospital of Jining Medical College, Shandong, 272029, People’s Republic of China; 3Department of Bioinformatics, The Basic Medical School of Chongqing Medical University, Chongqing 400016, People’s Republic of China; 4Department of Gastroenterology, Chongqing Public Health Medical Center, Chongqing, 400036, People’s Republic of China
Correspondence: Fangzhou Song
Molecular Medicine and Cancer Research Center, Chongqing Medical University, Yixueyuan Road, Yuzhong District, Chongqing 400016, People’s Republic of China
Tel +86 23 68485958
Email [email protected]
Purpose: Gastric cancer (GC) is the fifth most common tumor in the world, and most patients with GC have a poor prognosis. This study aimed to explore the biological influence and mechanism of LINC01272 in GC.
Materials and Methods: Using bioinformatic analyses, we investigated the expression of LINC01272 in TCGA database and predicted the biological functions and mechanism of LINC01272 in GC. Then, we detected the expression of LINC01272 in GC cell lines, GC tissues, and corresponding normal tissues using real-time polymerase chain reaction (RT-PCR). Finally, we explored the migration and invasion ability of LINC01272 by wound-healing and Transwell assays and examined the expression of epithelial-mesenchymal transition (EMT)-related proteins through Western blotting.
Results: We found that LINC01272 was upregulated in GC and was associated with GC staging and lymph node metastasis. The results of wound-healing and Transwell assays revealed that the LINC01272 was closely related to GC cell migration and invasion. LINC01272 knockdown inhibited the migration and invasion ability of GC cells by reducing the expression of EMT-related proteins. Overexpression of LINC01272 had the opposite effect.
Conclusion: Together, our results showed that LINC01272 promoted GC metastasis ability by regulating the expression of EMT-related proteins and could serve as a potential diagnostic biomarker for GC.
Keywords: gastric cancer, long non-coding RNA, migration, invasion, LINC01272
Introduction
Gastric cancer (GC) is one of the most common malignant tumors and the third leading cause of cancer mortality worldwide, with estimated 1,033,701 new cases and 7,82,685 deaths in 2018.1 More than 70% of new GC cases occur in developing countries, and the highest GC incidence and mortality have been reported in Eastern Asian countries, including China, Japan, and Korea.2,3 Most patients with GC are already at an advanced stage when diagnosed and have a poor prognosis, although chemotherapy and surgery are the main treatment options for advanced GC.4–6 One of the main reasons for the low survival rate of GC in 5 years is the lack of specific early diagnostic biomarkers. Therefore, it is of great clinical importance to explore the early diagnostic biomarkers of GC, which could increase diagnostic efficiency.7
Long non-coding RNA (lncRNA) is a type of an endogenous RNA that is longer than 200 nucleotides in length and has limited protein-coding potential. Dysregulated function or gene mutations in lncRNA cause a variety of human disease conditions, including birth defects and tumors, thus serving as a potential target for the treatment of certain diseases.8,9 However, the mechanism of lncRNA in human diseases is not clear.10 Emerging evidence indicates that lncRNA plays an important role in GC.11,12 For example, LINC00483,13 SOX2 overlapping transcript (SOX2OT),14 and ETS proto-oncogene 1 (Ets-1)15 have been reported to promote GC cell proliferation, apoptosis, migration, and invasion, and also serve as new biomarkers. Pvt1 oncogene (PVT1) can significantly induce angiogenesis, as well as promote tumor growth in vivo and in vitro in GC.16 By analyzing GC-related lncRNAs, we found that LINC01272 is closely related to the development of GC. In order to understand the functions of LINC01272 in GC, we investigated its expression levels in GC and its effects on the migration and invasion of GC cells. In conclusion, we found that LINC01272 plays an important role in GC cell migration and invasion and could serve as a novel diagnostic biomarker for GC.
Materials and Methods
Bioinformatics Analysis and Ethical Approval
The TCGA htseq-count expression data of stomach adenocarcinoma (n = 407, 375 tumors and 32 normal tissues) were downloaded from GDC data portal (https://portal.gdc.cancer.gov/repository). The clinical information of patients was also downloaded. The quantile normalized log2 count per million expression values were used to perform clustering analysis. The unsupervised hierarchical clustering was based on Euclidean distance metric with Ward linkage using pheatmap package in R software (version 3.6.1). Differential expression analysis was performed based on raw count data using DESeq2 package in R software to screen deregulated lincRNAs. LincRNAs with Benjamini-Hochberg adjusted P value <0.05 were considered statistically significant. To identify key functional lincRNAs in gastric cancer, the Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using DAVID online tools. The GEPIA online tool was used to compare lincRNA expression between tumor and normal tissues in diverse cancer types. Furthermore, the transcript per million (TPM) expressions of lincRNAs and genes were used to perform Pearson correlation analysis in GEPIA. 8 gastric tumor tissues and 5 normal stomach tissues were collected from patients with GC at the first affiliated hospital of Chongqing Medical University. We stated that all patients provided written informed consent. All experimental procedures were approved by the first affiliated hospital of Chongqing Medical University and Chongqing Medical University. We confirmed that all experiments were conducted in accordance with Declaration of Helsinki.
Cell Culture
The human GC cell lines, BGC823, SGC7901, N87, HGC-27, and AGS were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). GES-1 was purchased from Obio Technology Corp. (Shanghai, China). AGS was cultured in F12 medium and the other cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin. All cell lines were grown at 37°C in a humidified incubator with 5% CO2.
RNA Extraction and RT-PCR
Total RNA was extracted from tissues or cells using the Eastep® Super Total RNA Extraction Kit (Promega Biotechnology, Beijing, China), and reverse transcription was achieved with the PrimeScript™ RT Master Mix according to the manufacturer’s instructions (Takara Biotechnology, China). RT-PCR was performed using the SYBR Green PCR Kit (Bio-Rad, China). The primers used were as follows: LINC01272: 5ʹ-CCAAGGTCACGCAGCACAGTC-3ʹ and 5ʹ-GCAGAGATGAGCAGCAGTGGTG-3ʹ; β-actin: 5ʹ-TCCACGAGACCACCTACAAC-3ʹ and 5ʹ-TCTGGTGGGGCAATGATCTT-3ʹ. Primers against matrix metallopeptidase 9 (MMP-9), cadherin 2 (N-CAD), vimentin, Twist, and zinc finger E-box binding homeobox 2 (ZEB2) were designed and synthesized by Sangon Biotech (Shanghai, China). The PCR reaction conditions were as follows: 95°C for 5 min, followed by 39 cycles at 95°C for 10 s and 60°C for 30 s.
RNA Interference and Transfection
All small interfering RNA (siRNAs) used in this study were chemically synthesized by GenePhama (Shanghai, China). The siRNA sequences were as follows: si226: 5ʹ-GCAGCACAGUCACAUCCUATT-3ʹ and 5ʹ-UAGGAUGUGACUGUGCUGCTT-3ʹ; si342: 5ʹ-GCGCCUGUCCAGGACAAGUTT-3ʹ and 5ʹ-ACUUGUCCUGGACAGGCGCTT-3ʹ; si478: 5ʹ-GCACAGAAGUCUCUUCCCUTT-3ʹ and 5ʹ-AGGGAAGAGACUUCUGUGCTT-3ʹ. The negative control sequence was as follows: 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ and 5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ. Cells were transfected with RNAi MAX (Thermo Fisher Scientific, Waltham, MA, USA). At 48 h after transfection, cells were collected.
Amplification and Extraction of Plasmid
LINC01272 overexpression plasmid and control plasmid (pcDNA 3.1) were purchased from Sangon Biotech (Shanghai, China). The plasmid extraction kit was purchased from Takara Biotechnology. AGS cells were seeded in 6-well plates containing F12 medium and infected with the plasmid. At 48 h after infection, cells were collected.
Migration and Invasion Assays
Cell migration and invasion abilities were assessed by wound-healing and Transwell assays. For the wound-healing assay, about 4 × 105 cells were seeded into a 6-well plate. After transfection with siRNA for over 24 h, a 200 μL pipette tip was used to scrape cells. The cells were imaged under a microscope (Nikon, Japan) at different time points. The distance of the uncovered wound gap was quantified using Image J software. For the Transwell migration assay, about 4 × 104 cells infected with siRNA were resuspended in 200 μL serum-free medium and seeded into the upper chamber. Complete medium (700 μL) was added to the lower chamber as chemotactic factor. For the Transwell invasion assay, invasion was assessed based on the migration experiment. Cells were seeded onto a Matrigel-coated Transwell chamber. After 48 h, the cells that had migrated to the lower surface were fixed and stained with 0.5% crystal violet, and visualized and imaged under a microscope. Each experiment was performed at least three times.
Western Blotting
The treated cells were lysed in an appropriate volume of RIPA lysis buffer (Servicebio, China) containing 1% phenylmethanesulfonyl fluoride (Servicebio, China) and placed on ice for 30 min. Next, the mixture was centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was collected and protein concentration was determined by the bicinchoninic acid assay (Beyotime Biotechnology, Shanghai, China). Then, 5x loading buffer (Beyotime Biotechnology, Shanghai, China) was added to the supernatant and heated at 100°C for 15 min. The proteins were separated on sodium dodecyl sulfate-polyacrylamide gels (Beyotime Biotechnology, Shanghai, China) and transferred to polyvinylidene fluoride (PVDF) membranes (Merck Millipore). The membrane was blocked in 5% skimmed milk at room temperature for 2 h and then incubated with primary antibody (β-actin antibody was diluted 1:4000, other antibodies (MMP-9, N-CAD, Twist, Vimentin and ZEB2) were diluted 1:1000) overnight at 4°C, followed by incubation with appropriate secondary antibody for 2 h at room temperature. Protein expression was detected using the ECL kit (Advansta, San Jose, CA, USA). The primary antibodies (β-actin, MMP-9, N-CAD, Twist, Vimentin and ZEB2) and the secondary antibody were purchased from Servicebio.
Statistical Analysis
All in vitro experiments were repeated independently three times or more, and statistical data were analyzed by the SPSS statistical software (IBM Corporation, Armonk, NY, USA). All data were represented as mean ± standard deviation (SD), and Student’s t-test and one-way analysis of variance (ANOVA) were used for data analysis. P<0.05 was considered statistically significant.
Results
Analysis of LINC01272 by Bioinformatics Method
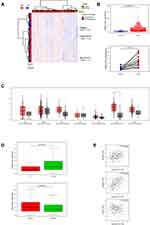
Read count data of TCGA-STAD were downloaded. A total of 375 GC tissues and 32 normal tissues were used for analysis. A heat map of hierarchical cluster analysis indicated that expression of genes differed between GC and normal tissues (Benjamini-Hochberg adjusted P value <0.05, and |logFC| > 1, Figure 1A). Analysis of the TCGA data revealed that LINC01272 expression was significantly higher in GC tissues than that in normal tissues. Moreover, in the paired samples, the expression of LINC01272 was significantly upregulated in GC tissues compared to that in adjacent normal tissues (P<0.001, Figure 1B). The results of GO function and KEGG pathway enrichment analyses showed that the main signaling pathways affected by LINC01272 were Toll-like receptor signaling pathway, NF-kappa B signaling pathway, and Natural killer cell-mediated cytotoxicity (Supplementary 1). It has been shown that the EMT transcription factors are tightly associated with NF-kappa B signaling pathway in several tumors: prostate cancer,17 renal carcinoma.18 And NF-kappaB is involved in the regulation of EMT genes in breast cancer cells.19 The expression levels of LINC01272 in different tumor types have been shown in Figure 1C. Red represents the expression level of LINC01272 in tumor samples and grey represents that in normal samples. Using GEPIA online database, we found that LINC01272 was highly expressed in GC, but not in the other six cancer types. Moreover, LINC01272 expression was closely associated with GC staging and lymph node metastasis (Figure 1D). Besides, correlation analysis revealed that LINC01272 expression was positively correlated with the expression of EMT-related proteins (matrix metallopeptidase 2 (MMP-2) and MMP-9), and negatively correlated with the expression of tight junction protein 1 (TJP1) (Figure 1E).
LINC01272 Expression Is Upregulated in GC
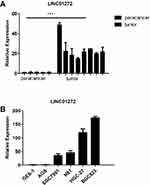
To further validate that LINC01272 expression is upregulated in GC, we measured the expression levels of LINC01272 in 8 GC tissues and 5 normal stomach tissues. The results of RT-PCR showed that the expression of LINC01272 was significantly high in GC tissues (P<0.05, Figure 2A). This is consistent with the high expression of LINC01272 in GC reported in TCGA database. In addition, LINC01272 expression was significantly higher in GC cell lines than that in normal gastric epithelial cell GES-1, and was the highest in the BGC823 cell line (lowest degree of differentiation), followed by HGC-27 cell line. (P<0.001, Figure 2B).
Knockdown of LINC01272 Inhibited GC Cell Migration and Invasion
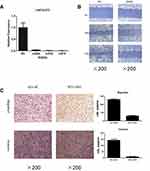
To explore the functions of LINC01272 in GC, we designed three siRNAs (si226, si342, and si478) targeting LINC01272 and transfected them into BGC823 cells. The expression levels of LINC01272 were examined by RT-PCR at 48 h post-transfection. RT-PCR results showed that transfection with si342 and si478 more effectively knocked down LINC01272 expression in BGC823 cells (Figure 3A). Therefore, we selected si342 for wound-healing and Transwell assays. The results of the wound-healing assay showed that the scratch width of BGC823 cells transfected with si342 was slightly altered, while the scratch disappeared in untreated BGC823 cells (Figure 3B). The results of Transwell assay showed that a significantly higher number of untreated BGC823 cells passed through the chamber compared to that in cells transfected with si342 (Figure 3C). These results suggested that knockdown of LINC01272 expression might inhibit the migration and invasion ability of BGC823 cells.
Overexpression LINC01272 Promoted Cell Migration and Invasion
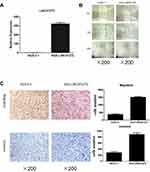
Due to their relatively low LINC01272 expression, AGS cells were chosen for LINC01272 overexpression. Forty-eight hours after transfection with LINC01272 overexpression plasmid, the expression level of LINC01272 was examined (Figure 4A). The results of wound-healing assay suggested that the metastatic ability of untreated AGS cells and cells transfected with overexpression plasmid was almost same at the beginning. However, at 12 and 24 h post-transfection, the metastasis of AGS cells transfected with overexpression plasmid was significantly increased compared to that of untreated AGS cells (Figure 4B). In addition, the number of untreated AGS cells that passed through the Transwell chamber significantly decreased compared to that of LINC01272-overexpressing cells (Figure 4C). These data indicated that the migration and invasion abilities of GC cells transfected with overexpression plasmid were significantly enhanced.
The Expression of Epithelial-Mesenchymal Transition (EMT)–Related Proteins
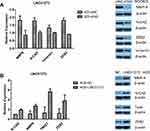
EMT occurs in the early stage of epithelial cell tumor metastasis, which is an important mechanism by which malignant tumor cells acquire migration and invasion abilities.20 It has been reported that tumors undergoing EMT-related processes become highly aggressive.21 To further verify the effect of LINC01272 on the migration and invasion of GC cells, we detected the expression of EMT-related transcription factors at the mRNA and protein levels using RT-PCR and Western blotting, respectively. The results revealed that knockdown of LINC01272 inhibited the expression of EMT-related transcription factors, including MMP-9, N-CAD, ZEB2, vimentin, and Twist (Figure 5A and B), while upregulation of LINC01272 showed the opposite results (Figure 5C and D).
Discussion
GC remains one of the most common malignancies worldwide, although there has been a decrease in both the incidence and mortality associated with GC in most developed countries in recent decades.1,2 The prognosis of several patients diagnosed with GC is poor, especially that of patients with advanced GC.22,23 Identification of specific early diagnostic markers is key to GC diagnosis. LncRNAs have been shown to be closely associated with lymph node metastasis, distant metastasis, and tumor differentiation in GC.24,25 In this study, we found that LINC01272 expression was upregulated in GC tissues and cells. Moreover, silencing of LINC01272 inhibited the migration and invasion ability of GC cells, while overexpression of LINC01272 had the opposite effect.
EMT refers to the biological process by which epithelial cells undergo specific changes to acquire a mesenchymal phenotype, and plays an important role in the development of human disease. Using proteomics, new molecular pathway related to this process has been identified including MAPK, PI3K/AKT, WNT pathways and Akt/mTOR signaling pathway, thus providing critical insights into the development of novel therapeutic targets.26,27 Recent studies have shown that EMT is associated with tumor cell migration and invasion, and cancer stem cell properties, including metastasis of glioma, human esophageal squamous cell carcinoma, and breast cancer.28–31 And Recent evidence indicated that some lncRNAs could regulate the EMT process to promote tumor development, for example, LINC00460 silencing inhibited EMT in colon cancer via Upregulating miR-433-3p,32 overexpression of GATA6 antisense RNA 1 (GATA6-AS1) Inhibited EMT through the Wnt/b-Catenin Signaling Pathway in GC,33 CHRF promoted GC cell invasion and migration via regulating EMT.34 Using bioinformatic analysis, we predicted that LINC01272 expression was associated with the expression of EMT-related proteins. We found that the expression of MMP-9, MMP-2, Twist, and ZEB2 were reduced in LINC01272 knockdown GC cells. Hence, we believe that LINC01272 promoted the metastasis of GC cells via reduction of the expression of MMP-2 and MMP-9.
Using a bioinformatics-based method, we found that LINC01272 was likely to be involved in GC Tumor Node Metastasis (TNM) stage. The expression level of LINC01272 in early GC was higher than that in advanced GC. We grouped N0 stage and N1 stage together. The expression level of LINC01272 might be upregulated in N1 stage compared to N0 stage, then with a decline in advanced N stage. In addition, the tumor metastasis is a complicated process, and LINC01272 could promote migration and invasion in early N stage. However, due to the insufficient GC patients’ clinical data, it is difficult to analyze the relationship between LINC01272 expression levels and N stage. Thus, it is still necessary to enlarge GC samples in our future study. Metastasis is the typical malignant behavior of GC and is the reason why GC is difficult to completely cure. Recent evidence have indicated that the expression of cancer susceptibility 19 (CASC19),35 Rho GTPase activating protein 27 pseudogene 1 (ARHGAP27P1),36 and small nucleolar RNA host gene 16 (SNHG16)37 is significantly associated with higher pathologic TNM stage and lymph node metastasis. However, the underlying mechanisms remain unclear and require further investigation.
In this study, our results indicated that LINC01272 is closely associated with migration and invasion in GC and may serve as a potential biomarker for the treatment of GC. Besides, LINC01272 is also highly expressed in other diseases. LINC01272 has been shown to be associated with a myeloid proinflammatory signature in Crohn’s disease and was significantly upregulated in inflammatory bowel disease.38,39 Moreover, LINC01272 is closely related to the survival time of lung squamous cell carcinoma.40 Nevertheless, the target genes are unidentified and the mechanism remains unclear. Further studies are required to investigate the mechanisms by which LINC01272 affects GC migration and invasion.
Conclusion
LINC01272, highly expressed in GC tissues, is closely associated with GC staging and lymph node metastasis. Moreover, LINC01272 enhances GC cell migration and invasion by regulating the expression of EMT-related proteins.
Acknowledgments
This study was funded by Key Research and Development of Social and People’s Livelihood (grant number cstc2018jscx-mszdX0031), Chongqing Basic Science and Frontier Technology Research Project (grant number cstc2018jcyjAX0199) and the Natural Science Foundation of Chongqing in China (grant number cstc2019jcyj-msxmX0259). We would like to thank all participants and the staff at Molecular Medicine and Cancer Research Center of Chongqing Medical University.
Author Contributions
Design and coordination: Xue Leng, Sen Wang and Fangzhou Song. Experiment performance and paper writing: Xue Leng. Data collection: Geli Liu. Bioinformatic analysis: Jing Song and Wanfeng Zhang. Data analysis: Xianqin Zhang. Paper revision: Li Rong and Yongping Ma. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflict of interest.
References
1. Ferlay JCM, Soerjomataram I, Mathers C, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi:10.1002/ijc.31937
2. Park JY, Forman D, Waskito LA, Yamaoka Y, Crabtree JE. Epidemiology of helicobacter pylori and CagA-positive infections and global variations in gastric cancer. Toxins. 2018;10(4):163. doi:10.3390/toxins10040163
3. Sugano K. Screening of gastric cancer in Asia. Best Pract Res Clin Gastroenterol. 2015;29(6):895–905. doi:10.1016/j.bpg.2015.09.013
4. Selim JH, Shaheen S, Sheu WC, Hsueh CT. Targeted and novel therapy in advanced gastric cancer. Exp Hematol Oncol. 2019;8(1):25. doi:10.1186/s40164-019-0149-6
5. Gamboa AC, Winer JH. Cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for gastric cancer. Cancers. 2019;11(11):1662. doi:10.3390/cancers11111662
6. Wang S, Liu Y, Feng Y, et al. A review on curability of cancers: more efforts for novel therapeutic options are needed. Cancers. 2019;11(11):1782. doi:10.3390/cancers11111782
7. Yoon H, Kim N. Diagnosis and management of high risk group for gastric cancer. Gut Liver. 2015;9(1):5–17. doi:10.5009/gnl14118
8. Yinxue Yang YD, Liu X, Cho WC. Advances in experimental medicine and biology. In: Crusio WE, Lambris JD, Rezaei N, editors Non-Coding RNAs in Colorectal Cancer. Vol 937: Springer; 2016.
9. Shuai Y, Ma Z, Lu J, Feng J. LncRNA SNHG15: a new budding star in human cancers. Cell Prolif. 2019;53:e12716.
10. Liang J, Wei X, Liu Z, et al. Long noncoding RNA CYTOR in cancer: a TCGA data review. Clin Chim Acta. 2018;483:227–233. doi:10.1016/j.cca.2018.05.010
11. Fattahi S, Kosari-Monfared M, Golpour M, et al. LncRNAs as potential diagnostic and prognostic biomarkers in gastric cancer: a novel approach to personalized medicine. J Cell Physiol. 2019;235:3189–3206.
12. Tianwen LXM, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612. doi:10.18632/oncotarget.6926
13. Li D, Yang M, Liao A, et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J Cell Mol Med. 2018;22:3875–3886.
14. Qu F, Cao P. Long noncoding RNA SOX2OT contributes to gastric cancer progression by sponging miR-194-5p from AKT2. Exp Cell Res. 2018;369(2):187–196. doi:10.1016/j.yexcr.2018.05.017
15. Li D, Chen Y, Mei H, et al. Ets-1 promoter-associated noncoding RNA regulates the NONO/ERG/Ets-1 axis to drive gastric cancer progression. Oncogene. 2018;37(35):4871–4886. doi:10.1038/s41388-018-0302-4
16. Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37(30):4094–4109. doi:10.1038/s41388-018-0250-z
17. Zhang Q, Helfand BT, Jang TL, et al. Nuclear factor-kappa B-mediated transforming growth factor-beta-induced expression of vimentin is an independent predictor of biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2009;15(10):3557–3567. doi:10.1158/1078-0432.CCR-08-1656
18. Pantuck AJ, An J, Liu H, Rettig MB. NF-kappaB-dependent plasticity of the epithelial to mesenchymal transition induced by Von Hippel-Lindau inactivation in renal cell carcinomas. Cancer Res. 2010;70(2):752–761. doi:10.1158/0008-5472.CAN-09-2211
19. Pires BR, Mencalha AL, Ferreira GM, et al. NF-kappaB is involved in the regulation of EMT genes in breast cancer cells. PLoS One. 2017;12(1):e0169622. doi:10.1371/journal.pone.0169622
20. Yue Q-Y, Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(4):1003–1010. doi:10.26355/eurrev_201802_14382
21. Sannino G, Marchetto A, Kirchner T, Grunewald TGP. Epithelial-to-mesenchymal and mesenchymal-to-epithelial transition in mesenchymal tumors: a paradox in sarcomas? Cancer Res. 2017;77(17):4556–4561. doi:10.1158/0008-5472.CAN-17-0032
22. Feng Q, May MT, Ingle S, Lu M, Yang Z, Tang J. Prognostic models for predicting overall survival in patients with primary gastric cancer: a systematic review. Biomed Res Int. 2019;2019:5634598. doi:10.1155/2019/5634598
23. Jiang YF, Zhang HY, Ke J, Shen H, Ou HB, Liu Y. Overexpression of LncRNA GHET1 predicts an unfavourable survival and clinical parameters of patients in various cancers. J Cell Mol Med. 2019;23(8):4891–4899. doi:10.1111/jcmm.14486
24. Li PF, Chen SC, Xia T, et al. Non-coding RNAs and gastric cancer. World J Gastroenterol. 2014;20(18):5411–5419. doi:10.3748/wjg.v20.i18.5411
25. Lin MT, Song HJ, Ding XY. Long non-coding RNAs involved in metastasis of gastric cancer. World J Gastroenterol. 2018;24(33):3724–3737. doi:10.3748/wjg.v24.i33.3724
26. Neagu M, Constantin C, Bostan M, et al. Proteomic technology “lens” for epithelial-mesenchymal transition process identification in oncology. Anal Cell Pathol. 2019;2019:3565970. doi:10.1155/2019/3565970
27. Yang J, Hou Y, Zhou M, et al. Twist induces epithelial-mesenchymal transition and cell motility in breast cancer via ITGB1-FAK/ILK signaling axis and its associated downstream network. Int J Biochem Cell Biol. 2016;71:62–71. doi:10.1016/j.biocel.2015.12.004
28. Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell. 2019;49(3):361–374. doi:10.1016/j.devcel.2019.04.010
29. Li ZG, Xiang WC, Shui SF, Han XW, Guo D, Yan L. 11 Long noncoding RNA UCA1 functions as miR-135a sponge to promote the epithelial to mesenchymal transition in glioma. J Cell Biochem. 2019.
30. Liu B, Li X, Li C, Xu R, Sun X. miR-25 mediates metastasis and epithelial-mesenchymal-transition in human esophageal squamous cell carcinoma via regulation of E-cadherin signaling. Bioengineered. 2019;10(1):679–688. doi:10.1080/21655979.2019.1687391
31. Demirkan B. The Roles of Epithelial-to-Mesenchymal Transition (EMT) and Mesenchymal-to-Epithelial Transition (MET) in breast cancer bone metastasis: potential targets for prevention and treatment. J Clin Med. 2013;2(4):264–282. doi:10.3390/jcm2040264
32. Hong W, Ying H, Lin F, Ding R, Wang W, Zhang M. lncRNA LINC00460 Silencing represses EMT in colon cancer through downregulation of ANXA2 via upregulating miR-433-3p. Mol Ther Nucleic Acids. 2020;19:1209–1218. doi:10.1016/j.omtn.2019.12.006
33. Li ZT, Zhang X, Wang DW, et al. Overexpressed lncRNA GATA6-AS1 inhibits LNM and EMT via FZD4 through the Wnt/β-catenin signaling pathway in GC. Mol Ther Nucleic Acids. 2020;19:827–840. doi:10.1016/j.omtn.2019.09.034
34. Gong J, Wang Y, Shu C. LncRNA CHRF promotes cell invasion and migration via EMT in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24(3):1168–1176. doi:10.26355/eurrev_202002_20168
35. Wen-Jie Wang C-AG, Rui L, Xu Z-P, et al. Long non-coding RNA CASC19 is associated with the progression and prognosis of advanced gastric cancer. Aging (Albany NY). 2019;11(15):5829–5847. doi:10.18632/aging.102190
36. Guohua Zhang YX, Zou C, Tang Y, et al. Long noncoding RNA ARHGAP27P1 inhibits gastric cancer cell proliferation and cell cycle progression through epigenetically regulating p15 and p16. Aging (Albany NY). 2019;11(20):9090–9110. doi:10.18632/aging.102377
37. Pang W, Zhai M, Wang Y, Li Z. Long noncoding RNA SNHG16 silencing inhibits the aggressiveness of gastric cancer via upregulation of microRNA-628-3p and consequent decrease of NRP1. Cancer Manag Res. 2019;11:7263–7277. doi:10.2147/CMAR.S211856
38. Haberman Y, BenShoshan M, Di Segni A, et al. Long ncRNA landscape in the ileum of treatment-naive early-onset crohn disease. Inflamm Bowel Dis. 2018;24(2):346–360. doi:10.1093/ibd/izx013
39. Wang S, Hou Y, Chen W, et al. KIF9AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease. Mol Med Rep. 2018;17(2):2195–2202. doi:10.3892/mmr.2017.8118
40. Wen-Jie Chen R-XT, He R-Q, Li D-Y, et al. Clinical roles of the aberrantly expressed lncRNAs in lung squamous cell carcinoma: a study based on RNA-sequencing and microarray data mining. Oncotarget. 2017;8(37):61282–61304.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.