Back to Journals » Cancer Management and Research » Volume 11
Linc01194 acts as an oncogene in colorectal carcinoma and is associated with poor survival outcome
Authors Wang X, Liu Z, Tong H, Peng H, Xian Z, Li L, Hu B, Xie S
Received 29 September 2018
Accepted for publication 31 December 2018
Published 22 March 2019 Volume 2019:11 Pages 2349—2362
DOI https://doi.org/10.2147/CMAR.S189189
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Xiaoxue Wang, Zhimin Liu, Hong Tong, Hui Peng, Zhenyu Xian, Li Li, Bang Hu, Shangkui Xie
Department of Proctology, The Sixth Affiliated Hospital of Sun Yat-Sen University, Guangzhou, People’s Republic of China
Background: The incidence of colorectal cancer ranks among the top three malignant tumors, attributing to more than 50,000 deaths in the United States every year. Survival rate is directly correlated with TNM stage at diagnosis, and identifying the molecules involved in the cancer development process will provide directions to better investigate the mechanisms of colorectal cancer.
Materials and methods: Bioinformatics analysis of differentially expressed long noncoding RNAs (lncRNAs), survival analysis, cell proliferation assay, migration assay, and Western blot analysis were performed.
Results: Fifty-one lncRNAs were identified between the early stage and late-stage groups. In the survival analysis, we found that Linc01194 is correlated with poor survival of colon cancer patients. In addition, by suppressing the expression of Linc01194 in colon cancer cell lines, cell proliferation and migration were inhibited. Western blot showed that N-cadherin and vimentin were downregulated, whereas E-cadherin was upregulated indicating that the process of epithelial–mesenchymal transition (EMT) was restrained.
Conclusion: Linc01194 promotes the proliferation and migration ability of colon cancer cells by activating EMT. It acts as an oncogene in colorectal carcinoma and is associated with worse survival outcome.
Keywords: colorectal cancer, TCGA, lncRNA, Linc01194, EMT
Introduction
The incidence of colorectal cancer ranks among the top three malignant tumors, attributing to more than 50,000 deaths in the United States every year.1,2 Survival rate correlates directly with TNM stage at diagnosis, and the 5-year survival rate sharply decreases from 90% at the early stage to 10% at the late stage.3 The molecular mechanism of colorectal cancer has been investigated well, but its development process remains unknown. Identifying the molecules involved therein will provide directions to better investigate the mechanisms of colorectal cancer.
Previous studies have revealed that long noncoding RNA (lncRNA) has a nonnegligible function in colorectal cancer.4,5 PVT1 is over expressed in colon cancer tissues compared to adjunct tissues and promotes cell proliferation and migration through the miR-30d-5p/runt-related transcription factor 2 (RUNX2) axis.6,7 In addition, lncTCF7 could activate the Wnt/β-catenin signaling pathway to promote metastasis in colorectal cancer.8,9 The Cancer Genome Atlas (TCGA) supplies genome information including sequence information, methylation data, copy number variations, and corresponding clinical parameters.10 Taking advantage of these data, the metastasis of colon cancer can be studied thoroughly to provide directions for future investigation.
Bioinformatics analysis identified 51 lncRNAs between the early stage and late-stage groups, and Kaplan–Meier analysis identified four lncRNAs with prognostic value in colorectal cancer. Combined with the above analysis, we hypothesized that Linc01194 functions as an oncogene in colon cancer. To verify our hypothesis, assays for cell proliferation, migration, and invasion were performed. In addition, epithelial–mesenchymal transition (EMT) markers were detected. These findings suggest that Linc01194 could promote the migration of colorectal cancer cells by activating EMT, providing new insights to further study the metastasis of colon cancer.
Materials and methods
Sequence data and clinical parameters
Sequence data and clinical parameters of colorectal cancer were acquired from the TCGA, which is a comprehensive public data bank for research purposes. In this data bank, 467 colorectal cancer samples including 268 stage I/II and 199 stage III/IV samples were acquired as of August 1, 2018 with the use of Genomic Data Commons tool according to the guidelines. In terms of clinical parameters, vital status, and survival days (days to death plus days to follow-up) of corresponding patients were collected.
Differentially expressed lncRNAs
lncRNA data were derived from sequence data based on CRCh38.87 mapping. lncRNA expression was normalized using the calcNormFactors function in the edgeR R package within and among samples.11 In addition, to avoid low-abundance lncRNA, lncRNAs with an average expression of less than one in all samples were excluded. To identify differentially expressed lncRNAs, sequence data from stage III/IV samples were compared with those of stage I/II samples. In this procedure, the edgeR R package was utilized again. Differentially expressed lncRNAs were defined with a cutoff of false discovery rate (FDR)-adjusted P<0.01 and |logFC| ≥ 1.5 (FC = fold change).
Survival analysis
To discover candidate lncRNAs with prognostic value, Kaplan–Meier analysis was conducted to compare the overall survival of the top ten differentially expressed lncRNAs. lncRNAs were divided into two groups: low expression (expression lower than median) and high expression (expression higher than median). Log-rank test was employed to identify key lncRNAs, and P<0.05 was considered statistically significant.
Cell culture
The colorectal cancer cell lines HT29, HCT116 SW620, and LoVo were purchased from the Cell Bank of Typical Culture Collection (Chinese Academy of Sciences, Shanghai, People’s Republic of China). All cells were grown in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS, 100 µg/mL streptomycin sulfate, and 100 U/mL penicillin. Cells were cultured in a humidified incubator at 37°C with 5% CO2.
Small interfering RNA (siRNA) transfection
HCT116 cells were transfected with siRNA targeting Linc01194 or negative control siRNA (Shanghai GenePharma Co., Ltd., Shanghai, People’s Republic of China) using X-treme GENE siRNA transfection reagent (Roche Applied Science, Penzbeg, Germany) according to the protocol. The siRNA sequences were as follows: si-Linc01194, ACAUAUUUACCCAUGUAGCAA, and GCUACAUGGGUAAAUAUGUGC.
RNA extraction
RNA was extracted following the protocol of TRIzol reagent. Briefly, cells at 80%–90% confluence were washed thrice with cold PBS. Then, the cells were lysed with cold TRIzol for 1 minute on ice. Chloroform was added into the lysate and left standing for 10 minutes at room temperature following centrifugation at 12,000 rpm at 4°C for 15 minutes. Thereafter, the transparent liquid was extracted carefully, mixed with an equal volume of isopropanol, and left standing for 10 minutes at room temperature followed by centrifugation at 12,000 rpm at 4°C for 15 minutes. After centrifugation, the supernatant was discarded and the precipitate was washed with diethy pyrocarbonate (DEPC) for 5 minutes. Then, the precipitate was dissolved in DEPC and reverse transcribed into complementary DNA (cDNA) using transcriptor first strand cDNA synthesis kit (Takara Bio Inc., Kusatsu, Japan).
Real-time (RT)-PCR
The cDNA was diluted at a ratio of 1:10, and the reaction system was constructed based on the protocol of the RT-PCR kit (Takara Bio Inc.) using the 7500 Fast RT-PCR system (Thermo Fisher Scientific). The primers of GAPDH were: forward, CCTGCACCACCAACTGCTTA and reverse, GGCCATCCACAGTCTTCTGAG. The primers of Linc01194 were: forward, AGACTGCTCTTGAGGCTGGAGT and reverse, AGGCTGAGGCTGGAGGATCTCT.
Cell proliferation assay
HCT116 cells transfected with negative control or si-Linc01194 were seeded into 96-well plates. The Cell Counting Kit-8 assay (Dojindo Laboratories Co. Ltd., Kumamoto, Japan) was used to assess the proliferative capacity of the cells according to the protocol provided by the manufacturer. The absorbance was detected using a microplate reader (MTX LAB SYSTEMS INC, Vienna, VR, USA) at 450 nm after 24, 48, 72, and 96 hours. The average absorbance in each group was calculated from six wells.
Cell migration and invasion assay
For the cell migration assay, HCT116 cells transfected with negative control or si-Linc01194 were seeded onto the upper chamber in medium containing 1% FBS, and full growth medium was loaded in the lower chamber. After 24 hours, the cells were washed and fixed in 4% paraformaldehyde for half an hour. Then the cells in the upper layer were removed carefully. The membranes were stained with crystal violet for approximately half an hour. Cell counts were performed on five random fields in each well. Cell invasion assay was performed following a similar procedure as the cell migration assay, except that the membrane was pre-coated with Matrigel.
Cell cycle assay
After transfecting with negative control or si-Linc01194 for 72 hours, HCT116 cells were harvested and fixed with 75% ethanol and resuspended in reagent A (cell cycle staining kit, Hangzhou Multi Sciences (Lianke) Biotech Co., Ltd., Hangzhou, People’s Republic of China) according to the protocol supplied. About half an hour later, cells were analyzed by flow cytometry. Each experiment was carried out thrice.
Western blot analysis
Proteins from HCT116 cells transfected with negative control or si-Linc01194 were extracted by RIPA buffer containing proteinase inhibitor cocktail and phosphatase inhibitor cocktail. The proteins were separated using 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes. After blocking with BSA for 2 hours, the membranes were incubated with primary antibodies at 4°C overnight followed by incubation with secondary antibody for 2 hours. After washing thrice with tris-buffered saline/Tween 20, the membranes were incubated with Super Signal West Pico chemiluminescent substrate and analyzed using the GeneGnome HR Image Capture System.
Gene ontology analysis
mRNAs that may be involved in the competing endogenous RNA (ceRNA) network of Linc01194 were acquired from the StarBase database.12 These mRNAs were sequenced in the Gene Ontology function enrichment analysis using the clusterProfiler R package downloaded from the Bioconductor website.13 Terms with P<0.05 were considered as statistically significant.
Statistical analysis
The correlation between Linc01194 expression and corresponding clinical parameters including outcome details were analyzed by SPSS software using chi-squared test. P<0.05 was considered statistically significant. Other statistical analyses were performed as stated above.
Results
Differentially expressed lncRNAs
A total of 7,469 lncRNAs were identified in colorectal cancer samples. Among them, after comparing the stage III/IV samples with stage I/II samples, 51 lncRNAs were defined as differentially expressed according to the FDR cutoff <0.01 and |logFC| ≥ 1.5, as shown in Figure 1 and Table S1. The expression levels of these differentially expressed lncRNAs are illustrated in Figure 2. The top ten differentially expressed lncRNAs were BX322234.2, Linc00400, Linc01419, FP671120.3, AC011840.1, AC105243.1, Linc01468, Linc01790, Linc02476, and Linc01194.
Survival analysis of lncRNAs
Kaplan–Meier analysis was performed on the top ten differentially expressed lncRNAs, revealing four lncRNAs with prognostic value. As shown in Figure 3, their high expression correlated directly with poor outcome of colorectal carcinoma patients.
Expression of Linc01194 in colorectal cancer cell lines
To elucidate the possible function of Linc01194, we detected the expression level of Linc01194 in four colorectal cancer cell lines: HT29, HCT116, SW620, and LoVo. Using RT-PCR, we found that Linc01194 is highly expressed in HCT116 cells. Then, we transfected HCT116 cells with Linc01194 siRNA to detect the interference efficiency. As shown in Figure 4, the inhibition rate of siRNA was greater than 70%. Thus, the siRNA sequence could be used for the subsequent experiments.
Linc01194 promoted the proliferation, migration, and invasion abilities of HCT116 cells
After HCT116 cells were transfected with Linc01194 siRNA, assays for proliferation, migration, and invasion were performed. As shown in Figure 5A, after 48 hours, the proliferation of HCT116 cells transfected with si-Linc01194 decreased significantly, and this phenomenon continued for another 48 hours. Then, the migration and invasion abilities were examined following the procedures mentioned above. As shown in Figure 5B and C, the number of migrating or invading cells were sharply decreased compared to that in the control group. As shown in Figure 5D, compared to the control group, the cells transfected with si-Linc01194 were arrested in G0/G1 phase.
Linc01194 promoted EMT in colorectal cancer
Western blot was performed on HCT116 cells treated with Linc01194 siRNA, using β-actin as the reference gene. In Figure 6, the expressions of N-cadherin and vimentin, which are markers of EMT, were decreased significantly, whereas E-cadherin expression was increased.
Correlation between Linc01194 and clinical parameters
Patients were ranked by Linc01194 expression from lowest to highest. The first quarter was defined as the low-expression group, whereas the last quarter was defined as the high-expression group. In addition, clinical parameters including age, gender, body mass index, location of carcinoma, TNM staging, histological type, neoplasm status, and risk level were divided into two groups. The criteria are shown in Table 1 in detail. To investigate the correlation between Linc01194 and clinical factors, the chi-squared test was performed using SPSS 23.0 software. As described in Table 2, the expression level of Linc01194 had a statistically significant correlation with stage (P<0.001), N stage (P<0.001), and neoplasm status (P<0.001). Patients with higher expression of Linc01194 seemed to have increased possibility of aggressive cancer stage, lymph node metastasis, and survival with tumor.
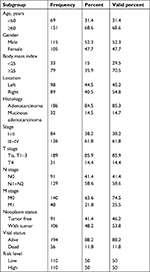  | Table 1 Clinical parameters of colorectal cancer patients |
  | Table 2 Relationship between Linc01194 expression and clinical parameters Note: *P<0.05. |
Gene Ontology analysis
As shown in Table S2, a total of 69 mRNAs that may be involved in the ceRNA network of Linc01194 were identified using the following criteria: common miRNA family ≥3, P-value <0.01, and FDR <0.01. Then, these genes were sequenced into the Gene Ontology function enrichment analysis. Of note, as shown in Table 3, only two terms, namely ubiquitin-protein transferase activity and ubiquitin-like protein transferase activity, were significantly enriched.
  | Table 3 Gene Ontology analysis of mRNA involved in the ceRNA network of Linc01194 Abbreviations: Bg ratio, background ratio; ceRNA, competing endogenous RNA. |
Discussion
lncRNAs are a type of noncoding RNAs that affect various biological processes, such as stemness maintenance, cell proliferation, and cell differentiation.14,15 Cancer is a complex disease, and lncRNAs play important roles in its progression and development.16,17 lncRNAs can exert the effects of ceRNA at cancer and act as either oncogenes or anti-oncogenes.18–20 In colon cancer research, it is important to identify biomarkers for early diagnosis and prognosis to provide recommendations for clinicians.
In order to explore the potential lncRNA biomarkers in colorectal cancer diagnosis and their associated biological functions, a bioinformatics assay was performed by comparing lncRNA sequencing data for the early stage (I and II) and late stage (III and IV) acquired from TCGA database. We found four lncRNAs, namely Linc01194, Linc00400, Linc02476, and AC011840.1, that were related to the prognosis of colorectal cancer. Among these, Linc00400 may have a relationship with coronary artery disease in the Japanese population.21 No research has reported the biological functions of the three other lncRNAs.
Cancer metastasis is closely related to cancer progression. In the first half of the investigation, Linc01194 was upregulated in late-stage colorectal carcinoma. Thus, we explored the biological function of Linc01194 by various methods. First, the expression levels of Linc01194 in HT29, HCT116, SW620, and LoVo colon cancer cells were evaluated, and Linc01194 expression was found to be the highest in HCT116 cells. Then, RNA interference experiments were performed in HCT116 cells to examine the impact of Linc01194 on colorectal cancer cells. Linc01194 knockdown had significant effects on the proliferation of colorectal cancer cells, which indicated that it may influence cell cycle progression. Besides fast proliferation, metastasis is another biological characteristic of cancer cells, and the highly metastatic characteristic of cancer are also correlated with poor prognosis. In our previous bioinformatics analysis, Linc01194 could be used as a biomarker to predict the prognosis of colorectal cancer patients, which indicated that there may be a correlation between Linc01194 and metastasis. In order to evaluate the biological effects of Linc01194 on metastasis, Transwell assays were performed to investigate cancer cell migration and invasion. By knocking down Linc01194, the number of cells moving through the membrane during the Transwell assays decreased significantly, which suggested that Linc01194 promoted cell movement. Transwell is a 3D cell culture method that can reliably simulate the in vivo microenvironment. The Transwell assays also showed that Linc01194 may greatly contribute to colon cancer metastasis. EMT is the central biological event that regulates metastasis,22,23 and it is necessary to explore the function of Linc01194 in EMT. E-cadherin, N-cadherin, and vimentin are the key markers in analyzing EMT. Linc01194 knockdown induced a decrease in the mesenchymal markers N-cadherin and vimentin, but increased the expression of E-cadherin, which is an epithelial marker. Moreover, lncRNAs can regulate microRNAs and decrease or increase the expression of genes via the ceRNA mechanism.24
Because Linc01194 is a relatively new lncRNA, we took advantage of databases to predict vital molecules influenced by Linc01194. Table S2 shows the mRNAs that may be involved in the ceRNA network of Linc01194. To investigate gene ontology in which these genes may be involved, we perform function enrichment analysis of these genes. Of note, GO:0004842 and GO:0019787 are involved in ubiquitin-protein transferase and ubiquitin-like protein transferase activity, respectively. These results indicated that changes in the ubiquitin system may be related to the progression and development of colorectal cancer. The ubiquitin system has complex biological functions and is a posttranslational modification that plays a role in several cellular processes including cell cycle progression, cell proliferation, DNA replication, and apoptosis.25,26 Research has shown that the ubiquitin system is frequently altered in various cancers and takes part in the vital movement of cancer, and can be used as a therapeutic target of cancer.27–29 Using bioinformatics, Linc01194 was predicted to upregulate the expression of BIRC6, BACH1, RNF2, etc, all of which play roles in the ubiquitination of proteins, via the ceRNA mechanism. BIRC6 is upregulated in various cancers, such as colon, liver, prostate, and lung cancer.30–33 In colon cancer, BIRC6 can be used as a biomarker for the prediction of prognosis, like Linc01194, which suggests that Linc01194 plays a central role in colon cancer. BACH1 is also an important gene regulated by Linc01194 that takes part in the development of breast, renal, colon, and ovarian cancers.34,35 It can induce the migration of colon cancer cells through reduction of metastasis-related genes. In our research, Linc01194 regulated the migration, invasion, and EMT progression of colon cancer cells. At the same time, Linc01194 may also regulate the expression of BACH1 through microRNA sponge function, indicating that Linc01194 may regulate colon cancer metastasis through the BACH1 gene. RNF2 is a type of RING-Type E3 ubiquitin transferase that is important for the function of the ubiquitination system and regulates cell cycle progression, apoptosis, DNA damage repair, and senescence.36 It is over expressed in many types of cancer, including melanoma, gastric, breast, and prostate cancers. However, the relationship between the RNF2 gene and colon cancer is unclear. Analysis of the ceRNA network gave us a promising direction, but the mechanisms of Linc01194 and the abovementioned genes need to be further investigated in future research.
Conclusion
In this study, we revealed that Linc01194 was over expressed in advanced colorectal cancer cells, predicting poor clinical outcome. As shown in Figure 7, Linc01194 induced proliferation and migration of colon cancer cells and regulated the expression of EMT-associated genes. The ceRNA network in which Linc01194 may be involved needs to be further investigated.
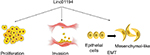  | Figure 7 The role Linc01194 plays in colorectal cancer. Notes: Linc01194 could promote proliferation, invasion, and EMT in colon cancer. Abbreviation: EMT, epithelial–mesenchymal transition. |
Acknowledgment
This study was supported by the Science and Technology Planning Project of Guangdong Province, People’s Republic of China (014A020212123).
Disclosure
The authors report no conflicts of interest in this work.
References
Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet. 2016;7(3):105–114. | ||
Price AS. Primary and secondary prevention of colorectal cancer. Gastroenterol Nurs. 2003;26(2):73–81. | ||
Moghimi-Dehkordi B, Safaee A. An overview of colorectal cancer survival rates and prognosis in Asia. World J Gastrointest Oncol. 2012;4(4):71–75. | ||
Xu MD, Qi P, Du X. Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Mod Pathol. 2014;27(10):24603586:1310–1320. | ||
Li H, Ma SQ, Huang J, Chen XP, Zhou HH. Roles of long noncoding RNAs in colorectal cancer metastasis. Oncotarget. 2017;8(24):28418892:39859–39876. | ||
Guo K, Yao J, Yu Q, et al. The expression pattern of long non-coding RNA PVT1 in tumor tissues and in extracellular vesicles of colorectal cancer correlates with cancer progression. Tumour Biol. 2017;39(4):101042831769912. | ||
Yu X, Zhao J, He Y. Long non-coding RNA PVT1 functions as an oncogene in human colon cancer through miR-30d-5p/RUNX2 axis. J Buon. 2018;23(1):48–54. | ||
Wu B, Chen M, Gao M, et al. Down-regulation of lncTCF7 inhibits cell migration and invasion in colorectal cancer via inhibiting TCF7 expression. Hum Cell. 2018;30225781. | ||
Li T, Zhu J, Wang X, et al. Long non-coding RNA lncTCF7 activates the Wnt/β-catenin pathway to promote metastasis and invasion in colorectal cancer. Oncol Lett. 2017;14(6):7384–7390. | ||
Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1A):A68–A77. | ||
Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. | ||
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. | ||
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J Integr Biol. 2012;16(5):284–287. | ||
Tordonato C, Di Fiore PP, Nicassio F. The role of non-coding RNAs in the regulation of stem cells and progenitors in the normal mammary gland and in breast tumors. Front Genet. 2015;6(e16915):72. | ||
Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. | ||
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. | ||
Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222. | ||
Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15(1):43. | ||
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. | ||
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. | ||
Yasukochi Y, Sakuma J, Takeuchi I, et al. Six novel susceptibility loci for coronary artery disease and cerebral infarction identified by longitudinal exome-wide association studies in a Japanese population. Biomed Rep. 2018;9(2):123–134. | ||
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. | ||
Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35(4):645–654. | ||
Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. | ||
Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12(9):1191–1197. | ||
Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422. | ||
Shi D, Grossman SR. Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol Ther. 2010;10(8):737–747. | ||
Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–444. | ||
Ding F, Xiao H, Wang M, Xie X, Hu F. The role of the ubiquitin-proteasome pathway in cancer development and treatment. Front Biosci. 2014;19:886–895. | ||
Hu T, Weng S, Tang W, et al. Overexpression of BIRC6 is a predictor of prognosis for colorectal cancer. PLoS One. 2015;10(5):e0125281. | ||
Tang W, Xue R, Weng S, et al. BIRC6 promotes hepatocellular carcinogenesis: interaction of BIRC6 with p53 facilitating p53 degradation. Int J Cancer. 2015;136(6):E475–E487. | ||
Luk SU, Xue H, Cheng H, et al. The BIRC6 gene as a novel target for therapy of prostate cancer: dual targeting of inhibitors of apoptosis. Oncotarget. 2014;5(16):6896–6908. | ||
Dong X, Lin D, Low C, et al. Elevated expression of BIRC6 protein in non-small-cell lung cancers is associated with cancer recurrence and chemoresistance. J Thorac Oncol. 2013;8(2):161–170. | ||
Liang Y, Wu H, Lei R, et al. Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J Biol Chem. 2012;287(40):33533–33544. | ||
Zhu GD, Liu F, Ouyang S, et al. BACH1 promotes the progression of human colorectal cancer through BACH1/CXCR4 pathway. Biochem Biophys Res Commun. 2018;499(2):120–127. | ||
Xia P, Wang S, Huang G, et al. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 2014;24(8):943–958. |
Supplementary materials
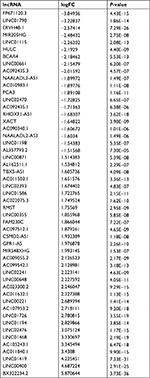  | Table S1 Fifty-one differently expressed lncRNAs in colon cancer Abbreviations: FC, fold change; lncRNAs, long noncoding RNAs. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.