Back to Journals » Cancer Management and Research » Volume 12
LINC01094 Down-Regulates miR-330-3p and Enhances the Expression of MSI1 to Promote the Progression of Glioma
Authors Zhu B, Liu W, Liu H, Xu Q, Xu W
Received 19 March 2020
Accepted for publication 23 June 2020
Published 28 July 2020 Volume 2020:12 Pages 6511—6521
DOI https://doi.org/10.2147/CMAR.S254630
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Bin Zhu,* Wei Liu,* Hongliang Liu, Qiang Xu, Wei Xu
Department of Neurosurgery, Huashan North Hospital, Baoshan Branch, Fudan University, Shanghai 200431, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Xu Email [email protected]
Background: This study aims at probing into the expression, function, and mechanism of LINC01094 and miR-330-3p in glioma.
Materials and Methods: qRT-PCR was employed to examine LINC01094 and miR-330-3p expressions in gliomas. After gain-of-function and loss-of-function models were constructed, CCK-8 and Transwell assays were used to detect the proliferation, migration and invasion of LN229 and U251 cells, respectively. Additionally, dual luciferase reporter gene assay was utilized to verify the binding site between m4iR-330-3p and LINC01094, miR-330-3p, and the 3ʹUTR of musashi RNA binding protein 1 (MSI1). Then, RNA pull-down, RIP, qRT-PCR and Western blot were employed to detect the regulatory relationships among LINC01094, miR-330-3p, and MSI1.
Results: The expression of LINC01094 was elevated in glioma tissues and cell lines, and the high expression of LINC01094 was associated with high grade of glioma. In contrast, miR-330-3p was lowly expressed in glioma tissue. Overexpression of LINC01094 or down-regulation of miR-330-3p promoted the proliferation, migration, and invasion of glioma cells, while LINC01094 knockdown or miR-330-3p up-regulation impeded these processes. miR-330-3p was identified as a target miRNA of LINC01094, and it could be negatively regulated by LINC01094. In addition, miR-330-3p antagonized the function of LINC01094 by negatively regulating MSI1.
Conclusion: LINC01094 promotes the proliferation, migration, and invasion of glioma cells by adsorbing miR-330-3p and up-regulating the expression of MSI1.
Keywords: LINC01094, miR-330-3p, MSI1, glioma
Introduction
Glioma, one of the deadliest malignancies, is challenging to be removed thoroughly in surgery, which is also not sensitive to radiotherapy and chemotherapy and is inclined to relapse.1–5 Therefore, it is of great significance to elucidate the molecular mechanism of glioma development and to find new molecular markers and targets for glioma.
Long non-coding RNA (lncRNA), or RNA molecule, does not encode proteins, whose length is longer than 200 nt.6 In the early days, lncRNA is regarded as the “waste” of transcription mediated by RNA polymerase II and has no biological effect.7 However, in recent years, it is confirmed that lncRNA participates in various biological processes.8 The abnormal expression of lncRNAs is closely related to various diseases including tumors.9,10 LINC01094 is proved to be highly expressed in clear cell renal cell carcinoma (ccRCC); knocking down LINC01094 inhibits the growth and metastasis of ccRCC cells.11 However, the expression, function and underlying mechanism of LINC01094 in gliomas are not clear.
MicroRNAs (miRNAs) can be fully or partially complementary to the untranslated region at the 3′ end of a specific mRNA and repress the process of translation.12 Differentially expressed miRNAs are closely linked to tumorigenesis and cancer progression.13 For example, miR-21 is highly expressed in glioma tissue and positively correlated with tumor grade.14 The expression of miR-221/222 in high-grade glioma tissues is higher than that in low-grade tumor tissues, and can directly target and regulate TIMP2 to modulate the tumor cell invasion.15 MircoRNA-330-3p (miR-330-3p) is an important member of miRNA, and its relationship with gliomas and the mechanism of action need to be studied.
Musashi RNA binding protein 1 (MSI1) is a highly conserved RNA-binding protein that is mainly expressed in the nervous system, controlling the balance between self-renewal and differentiation of neurons, and is regarded as a key oncogenic factor.16 MSI1 is highly expressed in many cancers, whose role is to facilitate tumor cell growth and suppress apoptosis.17–19 Previous researches validate that the expression of MSI1 in medulloblastoma tissue is higher than that in normal brain tissues, and its high expression is also related to the poor prognosis of glioma and medulloblastoma.20–23 These findings suggest that MSI1 promotes tumorigenesis and progression of gliomas.
Herein, we investigated the role of LINC01094 and miR-330-3p in the pathogenesis of gliomas and the regulatory mechanism of LINC01094/miR-330-3p/MSI1 axis in gliomas.
Materials and Methods
Clinical Samples and Ethics Statement
Tumor specimens were randomly collected from glioma patients who received surgery in Huashan Hospital from 2014 to 2018. Normal tissues and tumor tissues from 43 patients were immediately frozen in liquid nitrogen and kept at −196°C for the following experiments. Equipped with written informed consents signed by all patients involved in this study and supported by the Research Ethics Committee of Huashan Hospital, Fudan University, this study was carried out according to the Declaration of Helsinki.
Cell Lines and Cell Culture
Normal human astrocytes (NHA) cells and human glioma cell lines (U87, SHG-44, U251, LN229, and U373 cells) were obtained from the BeNa Culture Collection (BNCC, Beijing, China). NHA cells were cultivated in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, Carlsbad, CA, USA) supplemented with 25 mg/mL bovine insulin, 20 ng/mL epidermal growth factor, 20 ng/mL progesterone, and 50 mg/mL transferrin (Sigma, St. Louis, MO, USA). Human glioma cell lines were cultured in DMEM (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA). All cells were cultured in 5% CO2 and 90% relative humidity at 37°C.
Cell Transfection
Glioma cells were inoculated in a 24-well plate (2 × 105 cells/well) and cultured at 37°C in 5% CO2. With the cells’ confluence growing to 70%, the cells were transfected, which was performed employing Lipofectamine®3000 (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s protocol. LINC01094 overexpression plasmid, LINC01094 siRNAs, MSI1 overexpression plasmid, MSI1 siRNA, miR-330-3p mimics, anti-miR-330-3p, and their corresponding controls were available from GenePharma (Shanghai, China); 36 h after transfection, quantitative real-time polymerase chain reaction (qRT-PCR) was used to examine the transfection efficiency.
qRT-PCR
TRIzol reagent (Invitrogen, Shanghai, China) was used to extract total RNA from tissues and cells. cDNA was synthesized by reverse transcription employing PrimeScript™ RT kit (Takara, Dalian, China). RT-PCR was performed on ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) utilizing the SYBR Premix Ex Taq ™ Kit (Takara, Dalian, China). Besides, TaqMan-miRNA analysis (ThermoFisher Scientific, Waltham, MA, USA) was adopted to detect miR-330-3p expression. GAPDH and U6 small nuclear RNA were treated as endogenous controls for LINC01094, MSI1, and miR-330-3p, respectively. The relative expressions of LINC01094, miR-330-3p, and MSI1 were calculated by the 2-ΔΔCt method.
Cell Counting Kit-8 (CCK-8) Assay
Cell growth was measured with CCK-8 assay. Cells were inoculated in a 96-well plate (Corning Costar, Corning, NY) at a density of 10 × 103/well. After the cells were cultured for 24 h, 48 h, and 72 h, respectively, 10 μL CCK-8 solution (Beyotime, Shanghai, China) was dripped into each well in compliance with the protocol; 1 h later, the cell viability was observed with the absorbance of 450 nm; 72 h later, the proliferation curve was plotted.
Transwell Assay
The transfected cells were collected and the cell concentration was modulated to 1 × 105 cells/mL with serum-free medium; 200 μL the cell suspension of each group and 600 μL medium (containing 10% FBS) were, respectively, added to the upper chamber and the lower chamber of the Transwell chamber (8 μL pore size, Corning, Beijing, China) covered with Matrigel; 24 h later, after the chamber was removed, the cells remaining on the upper surface of the membrane were wiped off with cotton swabs, and those which passed through the membrane were fixed with paraformaldehyde, and then stained with crystal violet solution. After that, the cells in 5 randomly selected visual fields were counted under a microscope, and the average was taken to evaluate the invasion of the cells. There was no need for Matrigel coating in Transwell chamber in migration experiments, and the other steps followed the invasion assay.
Dual Luciferase Reporter Gene
Dual luciferase reporter plasmids of wild-type LINC01094-WT, MSI1-WT, mutant LINC01094-MUT, and MSI1-MUT were constructed. Then, LN229 and U251 cells in the logarithmic growth phase were inoculated into 12-well cell plates at 1 × 105 per well, followed by the co-transfection of LINC01094-WT/MSI1-WT, LINC01094-MUT/MSI1-MUT with miR-330-3p mimics or negative control into LN229 and U251 cells; 48 h later, the luciferase activity was detected using a dual-luciferase reporting system (Promega, Madison, WI, USA) in line with the protocol. The ratio of firefly luciferase activity to renilla luciferase activity was used to evaluate the binding between reporter plasmid and the miRNA.
RNA Immunoprecipitation (RIP) Assay and RNA Pull-Down Assay
For RIP assay, EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA) was used, and RIP analysis was performed according to the manufacturer’s protocol. In brief, glioma cells were lysed in lysis buffer with protease and RNase inhibitors. Subsequently, the protein extract was incubated with a washing buffer. Next, the RIP buffer contained magnetic beads conjugated with human anti-argonaute 2 (Ago2) antibody or mouse IgG was employed to incubate the extracts. After that, immunoprecipitation complex was incubated with proteinase K, and co-immunoprecipitated RNA was extracted. Moreover, qRT-PCR was performed to determine the enrichment of the targets.
For RNA pull-down assay, biotinylated miR-330-3p or negative control (NC) probes were conjugated with streptavidin beads (Beyotime, Shanghai, China). The probes were then transfected into the cells. The cells were incubated with the lysis buffer prior to the incubation of cell lysates with RNA-bound beads at 4°C for 2 h. Then the abundance of LINC01094 was analyzed by qRT-PCR.
Western Blot
RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) was utilized to extract total protein of cells. After being centrifuged, the supernatant was collected and then mixed with loading buffer. The protein was boiled for denaturation before the protein samples were subject to 12% SDS-PAGE. Subsequently, the protein was electrically transferred into the nitrocellulose (NC) membrane (Millipore, Bedford, MA, USA). The NC membrane was blocked with blocking solution (50mg/mL skim milk) for 2 h. After that, the primary antibodies anti-MSI1 (abcam, ab52865, 1:1000) and anti-GAPDH (abcam, ab181602, 1:1000) were employed to incubate the NC membrane at 4°C overnight. Afterward, the NC membrane was immersed in TBST for 10 min for 3 times, and horseradish peroxidase-labeled secondary antibody (abcam, ab205718, 1:2000) was loaded and the NC membrane was incubated at room temperature for 1 h. Then the membrane was rinsed with TBST for 3 times again. Ultimately, ECL chemiluminescent solution (Beyotime Biotechnology, Shanghai, China) was used to show the bands.
Statistical Processing
Data analysis was performed employing SPSS 21.0 (SPSS Inc., Chicago, IL, USA). Data were expressed as x ± s. The statistical analysis was carried out by t-test. P < 0.05 signified statistical significance.
Results
LINC01094 Expression Was Up-Regulated in Glioma, Which Was Related to Glioma Grading
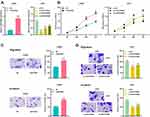
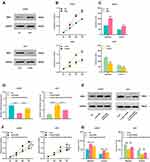
In order to investigate the expression of LINC01094 in gliomas, we adopted the GEPIA database to perform bioinformatics analysis, and it was found that LINC01094 was differentially expressed in grade IV glioma (glioblastoma, GBM) and normal brain tissues, and the expression in GBM tissues was considerably higher than in normal tissues (Figure 1A). To confirm this, we used qRT-PCR to probe LINC01094 expressions in cancer tissues and adjacent normal tissues in 43 glioma patients, and the results depicted that LINC01094 expression was remarkably up-regulated in 23 cases of grade Ⅰ-Ⅱ glioma tissue and 20 cases of grade Ⅰ-Ⅱ glioma tissue compared with adjacent cancer tissues; in contrast to low-grade glioma samples (Glioma Ⅰ-Ⅱ), the expression of LINC01094 was dramatically increased in high-grade glioma samples (Glioma Ⅰ-Ⅱ) (Figure 1B). In addition, we also tested LINC01094 expression in five glioma cell lines (U87, SHG-44, U251, LN229, and U373 cells), and the results implied that the expression of LINC01094 in all five glioma cell lines was markedly elevated in contrast to normal cell line NHA. These results suggested that LINC01094 was highly expressed in glioma and was positively correlated with high grade of glioma.
LINC01094 Promoted Proliferation, Migration and Invasion of Glioma Cells
According to the above results, in glioma cell lines, the lowest and the highest expression of LINC01094 existed in LN229 cells and U251 cells, respectively. Therefore, LN229 and U251 cell lines were selected as the cell models for further experiments. We transfected LNC01094 overexpressing plasmid into LN229 cells to construct a cell model of LINC01094 overexpression. Three LINC01094 siRNAs were transfected into U251 cells to construct knockdown models and si-LINC01094#1 and si-LINC01094#2 were selected for experiment (Figure 2A). Next, CCK-8 experiment was performed, the result of which showed that overexpression of LINC01094 significantly promoted the proliferation of LN229 cells, while knocking down LINC01094 significantly restrained the proliferation of U251 cells (Figure 2B). Additionally, Transwell experiments revealed that LINC01094 overexpression markedly enhanced the migration and invasion of LN229 cells, while knocking down LINC01094 observably reduced the migration and invasion of U251 cells (Figure 2C and D).
LINC01094 Can Function as a Molecular Sponge for miR-330-3p
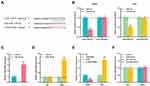
Bioinformatics tool StarBase v3.0 revealed the potential binding site between LINC01094 and miR-330-3p (Figure 3A). To verify whether LINC01094 could target miR-330-3p, we co-transfected wild-type luciferase reporter plasmid (LINC01094-WT) or mutant luciferase reporter plasmid (LINC01094-MUT) with miR-330-3p mimics or control miRNA into LN229 and U251 cells, respectively. As was shown, miR-330-3p mimics significantly reduced the luciferase activity of the LINC01094-WT reporter plasmid, while the luciferase activity of LINC01094-MUT reporter plasmid was not markedly changed (Figure 3B). Furthermore, RNA pull-down and RIP experiments proved that LINC01094 could directly interact with miR-330-3p (Figure 3C and D). In addition, qRT-PCR results displayed that miR-330-3p expression in LN229 cells was dramatically constrained after overexpression of LINC01094; miR-330-3p expression was increased in U251 cells after knocking down LINC01094 (Figure 3E). Moreover, miR-con/miR-330-3p or anti-miR-con/anti-miR-330-3p were transfected into U251 and LN229 cells, respectively, and as shown, LINC01094 expression was not changed by miR-330-3p (Figure 3F). In summary, these data suggested that LINC01094 adsorbed miR-330-3p and negatively modulated its expression in glioma cells.
miR-330-3p Impeded the Malignant Biological Behavior of Glioma Cells
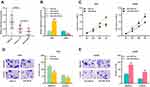
Next, we investigated miR-330-3p expressions in glioma tissues. The results showed that compared with adjacent cancer tissues, miR-330-3p expressions were significantly reduced in Ⅰ-Ⅱ grade glioma tissues and Ⅰ-Ⅱ glioma tissues; in glioma tissues, miR-330-3p expressions were observably lower in high-level glioma samples (Glioma Ⅰ-Ⅱ) than that in low-level glioma samples (Glioma Ⅰ-Ⅱ) (Figure 4A). U251 and LN229 cells were then transfected with miR-330-3p mimics and inhibitors, respectively, and qRT-PCR results confirmed that the transfection was successful (Figure 4B). Then the proliferation and metastasis of cells were evaluated by CCK-8 and Transwell experiments, respectively. It was observed that up-regulating miR-330-3p repressed the proliferation and metastasis ability of U251 cells (Figure 4C and D). Besides, in LN229 cells, down-regulating miR-330-3p exerted the opposite effect (Figure 4C and E). These results implied that miR-330-3p inhibited the malignant biological behaviors of glioma cells.
MSI1 Was the Target of miR-330-3p
StarBase v3.0 also predicted a complementary binding site between miR-330-3p and the 3ʹUTR of MSI1 (Figure 5A). To validate whether miR-330-3p could bind to the 3ʹUTR of MSI1, MSI1-WT reporter plasmid and MSI1-MUT reporter plasmid were co-transfected with miR-330-3p mimics or control miRNAs into U251 and LN229 cells, respectively. Dual luciferase reporter assay showed that miR-330-3p mimics can markedly reduce the luciferase activity of the MSI1-WT reporter plasmid compared with the control group, and there was no significant change in the luciferase activity of MSI1-MUT reporter plasmid (Figure 5B). Additionally, it was discovered that up-regulating miR-330-3p induced a significant decrease in the expression of MSI1 in U251 cells, and after inhibiting miR-330-3p, the expression of MSI1 in LN229 cells was increased (Figure 5C and D).
LINC01094 Regulated Glioma Cell Proliferation and Metastasis Through miR-330-3p/MSI1 Axis
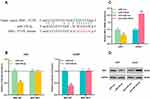
Next, we transfected MSI1 overexpressing plasmid into LN229 cells and MSI1 siRNA into U251 cells to construct a cell model of MSI1 overexpression or knockdown (Figure 6A). Western blot was employed to detect cell migration and invasion and it was found that MSI1 promoted glioma cell migration and invasion (Figure 6B and C). To further explore the mechanism between miR-330-3p and LINC01094, miR-330-3p mimics were transfected into LN229 cells overexpressing LINC01094, and anti-miR-330-3p was transfected into U251 cells with LINC01094 knockdown. qRT-PCR verified the success of transfection (Figure 6D). After detecting the protein level of MSI1 by Western blot, it was found that up-regulation of miR-330-3p partially reduced the increase in MSI1 protein expression resulted from LINC01094 overexpression, while antagonism of miR-330-3p partially reversed the decrease in MSI1 protein caused by LINC01094 knockdown (Figure 6E). In addition, functional experiments showed that the effect of LINC01094 overexpression on proliferation and metastasis of LN229 cells was partially weakened by miR-330-3p mimics; additionally, the inhibition of U251 cell proliferation and metastasis resulted from LINC01094 knockdown was partially reversed by anti-miR-330-3p (Figure 6F and G). Above findings indicated that LINC01094 was involved in regulating the malignant biological behaviors of glioma cells through regulating the miR-330-3p/MSI1 axis.
Discussion
Glioma, the most common primary brain tumors, accounts for about 80% of primary malignant brain tumors.24,25 It is believed that the molecular pathogenesis of glioma is accompanied with various genetic changes that lead to abnormal cell proliferation, cell movement, angiogenesis, cell cycle regulation, and apoptosis.26 According to its histological characteristics, the World Health Organization (WHO) divides brain glioma into four grades: grade I to IV, and the malignancy increases with the rise of grade.27 Although a lot of progresses have been made in surgery, radiotherapy, and chemotherapy in recent years, the therapeutic effect is still unsatisfactory due to tumor invasion and high recurrence rate.28
In recent years, a large number of lncRNAs are discovered as new regulatory molecules in cancer biology.29,30 For example, lncRNA UCA1 is upregulated in gliomas and promotes cancer cell proliferation.31 LncRNA ANRIL is overexpressed during glioma development and is associated with unfavorable pathological characteristics.32 In this study, it was demonstrated that LINC01094 was highly expressed in glioma tissues and cells and positively correlated with high grade of glioma. Knockdown of LINC01094 impeded the proliferation, migration, and invasion of glioma cells, while overexpression of LINC01094 facilitated the aforementioned biological behaviors of glioma cells. Therefore, for the first time, it was concluded that LINC01094 played a crucial role in glioma progression.
A single miRNA can modulate the expressions of multiple genes and vital biological processes (such as cell proliferation, differentiation, migration, as well as apoptosis) in cancer progression.33,34 miR-330-3p has distinct functions in different tumors. In breast cancer, miR-330-3p is highly expressed in cancer tissues and boosts cancer metastasis by targeting CCBE1.35 However, in gastric cancer, miR-330-3p is lowly expressed in cancer tissue and cells, and overexpression of miR-330-3p can inhibit the cancer progression by targeting MSI1.36 In addition, miR-330-3p restrains the migration of liver cancer cells by targeting MAP2K1.37 The expression and function of miR-330-3p in glioma are not reported previously. In this study, it was demonstrated for the first time that miR-330-3p was reduced in glioma tissues compared with normal brain tissues. Ectogenic miR-330-3p inhibited the proliferation, migration, and invasion of glioma cells, while inhibiting miR-330-3p functioned oppositely. Our data proved for the first time that miR-330-3p served as a tumor-suppressive miRNA in gliomas.
In recent years, competitive endogenous RNA (ceRNA) is a hot topic in cancer research. In this mechanism, lncRNA can regulate the translation of mRNA by competitively sponging miRNA via miRNA response elements (MREs).38 So far, accumulating lncRNAs have been identified as ceRNA in gliomas. For example, lncRNA MALAT1 can regulate the expression of SOX2 by sponging and repressing miR-129 in gliomas.39 LncRNA FTH1P3 promotes glioma progression by working as a molecular sponge for miR-224-5p.40 In this work, we hypothesized that LINC01094 could function as a molecular sponge to regulate miRNA in gliomas. Based on bioinformatics analysis and validation experiments, this study demonstrated that LINC01094 was a molecular sponge for miR-330-3p in gliomas. Subsequently, we proved that miR-330-5p partially reversed the effects of LINC01094 overexpression on glioma proliferation, migration, and invasion. These data indicated that LINC01094 could serve as ceRNA to participate in glioma progression via suppressing miR-330-3p.
MSI1 belongs to the Musashi family and is a marker gene for the progenitor maintenance and differentiation of neurons.41 Musashi members are abnormally expressed in a variety of tumor cells, and contribute to the malignant biological behaviors of cancer cells.42 The overexpression of MSI1 in cancer cells activates multiple oncogenic signaling pathways. In cervical cancer, it is verified as a promoter of epithelial-mesenchymal transition of cancer cells.16 It activates the STAT3 signaling to facilitate the progression of oral squamous cell carcinoma.17 In glioma, MSI1 up-regulates intercellular adhesion molecule-1, down-regulates Tensin3, and activates PKR/eIF2α signaling and IL-6/Akt signaling to mediate the migration and invasion of glioma cells, and its overexpression is linked to the chemoresistance and radioresistance of glioma cells.21,42–45 However, there are few researches focusing on the mechanism of MSI1 dysregulation in glioma. In the present work, it was found that MSI1 was a downstream target of miR-330-3p in gliomas, which was consistent with the study on gastric cancer.36 Additionally, it was also confirmed that LINC01094 positively regulated the expression of MSI1 through suppressing miR-330-3p. Our data indicated that the dysregulation of LINC01094/miR-330-3p axis was responsible for the abnormal expression of MSI1 in glioma.
In summary, we demonstrate for the first time that LINC01094 expression is elevated in gliomas, which enhances the proliferation, migration, and invasion of glioma cells. In terms of mechanism, LINC01094 promotes the malignant biological behaviors of glioma cells by functioning as a molecular sponge for miR-330-3p and indirectly up-regulating MSI1 expression. Thus, LINC01094 is a potential promising target for glioma treatment.
Author Contributions
Bin Zhu and Wei Xu designed the study. Bin Zhu did literature and experimental research, analyzed data, and prepared and edited the manuscript. Wei Xu reviewed the manuscript. Bin Zhu and Wei Liu finished statistical analysis. Hongliang Liu and Qiang Xu did clinical research and accomplished data acquisition. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors reported no conflicts of interest in this work.
References
1. Wu P, Cai J, Chen Q, et al. Lnc-TALC promotes O6-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat Commun. 2019;10(1):2045. doi:10.1038/s41467-019-10025-2
2. Mughal AA, Zhang L, Fayzullin A, et al. Patterns of invasive growth in malignant gliomas-the hippocampus emerges as an invasion-spared brain region. Neoplasia. 2018;20(7):643–656. doi:10.1016/j.neo.2018.04.001
3. Duffau H. Surgery of insular gliomas. Prog Neurol Surg. 2018;30:173–185. doi:10.1159/000464393
4. Afshari AR, Mollazadeh H, Mohtashami E, et al. Protective role of natural products in glioblastoma multiforme: a focus on nitric oxide pathway. Curr Med Chem. 2020;27. doi:10.2174/0929867327666200130104757
5. Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. doi:10.1186/s12943-018-0812-2
6. Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem. 2017;17(15):1750–1757. doi:10.2174/1568026617666161116144744
7. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi:10.3390/ijms19051310
8. Rajagopal T, Talluri S, Akshaya RL, Dunna NR. HOTAIR LncRNA: a novel oncogenic propellant in human cancer. Clin Chim Acta. 2020;503:1–18. doi:10.1016/j.cca.2019.12.028
9. Goyal N, Kesharwani D, Datta M. Lnc-ing non-coding RNAs with metabolism and diabetes: roles of lncRNAs. Cell Mol Life Sci. 2018;75(10):1827–1837. doi:10.1007/s00018-018-2760-9
10. Bao MH, Szeto V, Yang BB, Zhu SZ, Sun HS, Feng ZP. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018;9(3):281. doi:10.1038/s41419-018-0282-x
11. Jiang Y, Zhang H, Li W, Yan Y, Yao X, Gu W. FOXM1-activated LINC01094 promotes clear cell renal cell carcinoma development via microRNA 224-5p/CHSY1. Mol Cell Biol. 2020;40(3). doi:10.1128/MCB.00048-20
12. Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20(1):21–37. doi:10.1038/s41580-018-0045-7
13. Qadir MI, Faheem A. miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27(3):197–204. doi:10.1615/CritRevEukaryotGeneExpr.2017019494
14. Masoudi MS, Mehrabian E, Mirzaei H. MiR-21: a key player in glioblastoma pathogenesis. J Cell Biochem. 2018;119(2):1285–1290. doi:10.1002/jcb.26300
15. Yang F, Wang W, Zhou C, et al. MiR-221/222 promote human glioma cell invasion and angiogenesis by targeting TIMP2. Tumour Biol. 2015;36(5):3763–3773. doi:10.1007/s13277-014-3017-3
16. Gong P, Wang Y, Gao Y, et al. Msi1 promotes tumor progression by epithelial-to-mesenchymal transition in cervical cancer. Hum Pathol. 2017;65:53–61. doi:10.1016/j.humpath.2016.12.026
17. Wang CF, Zhang HC, Feng XM, Song XM, Wu YN. Knockdown of MSI1 inhibits the proliferation of human oral squamous cell carcinoma by inactivating STAT3 signaling. Int J Mol Med. 2019;44(1):115–124. doi:10.3892/ijmm.2019.4181
18. Liu L, Qiu F, Chen J, et al. Functional polymorphism in the MSI1 gene promoter confers a decreased risk of lung cancer in Chinese by reducing MSI1 expression. Curr Genomics. 2018;19(5):375–383. doi:10.2174/1389202919666171128151544
19. Niu J, Zhao X, Liu Q, Yang J. Knockdown of MSI1 inhibited the cell proliferation of human osteosarcoma cells by targeting p21 and p27. Oncol Lett. 2017;14(5):5271–5278. doi:10.3892/ol.2017.6870
20. Vo DT, Subramaniam D, Remke M, et al. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am J Pathol. 2012;181(5):1762–1772. doi:10.1016/j.ajpath.2012.07.031
21. Lin JC, Tsai JT, Chao TY, Ma HI, Liu WH. Musashi-1 enhances glioblastoma migration by promoting ICAM1 translation. Neoplasia. 2019;21(5):459–468. doi:10.1016/j.neo.2019.02.006
22. Yi C, Li G, Ivanov DN, et al. Luteolin inhibits Musashi1 binding to RNA and disrupts cancer phenotypes in glioblastoma cells. RNA Biol. 2018;15(11):1420–1432. doi:10.1080/15476286.2018.1539607
23. Lin JC, Tsai JT, Chao TY, Ma HI, Chien CS, Liu WH. MSI1 associates glioblastoma radioresistance via homologous recombination repair, tumor invasion and cancer stem-like cell properties. Radiother Oncol. 2018;129(2):352–363. doi:10.1016/j.radonc.2018.09.014
24. Chen Y, Zhao F, Cui D, et al. HOXD-AS1/miR-130a sponge regulates glioma development by targeting E2F8. Int J Cancer. 2018;142(11):2313–2322. doi:10.1002/ijc.31262
25. Du S, Yang S, Zhao X, et al. Clinical characteristics and outcome of children with relapsed medulloblastoma: a retrospective study at a single center in China. J Pediatr Hematol Oncol. 2018;40(8):598–604. doi:10.1097/MPH.0000000000001241
26. Soukhtanloo M, Mohtashami E, Maghrouni A, et al. Natural products as promising targets in glioblastoma multiforme: a focus on NF-κB signaling pathway. Pharmacol Rep. 2020;72(2):285–295. doi:10.1007/s43440-020-00081-7
27. Colip C, Oztek MA, Lo S, Yuh W, Fink J. Updates in the neuoroimaging and WHO classification of primary CNS gliomas: a review of current terminology, diagnosis, and clinical relevance from a radiologic prospective. Top Magn Reson Imaging. 2019;28(2):73–84. doi:10.1097/RMR.0000000000000195
28. Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of miR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–1157. doi:10.1007/s13311-018-0649-9
29. Zhang Z, Liu J, Wu Y, et al. Long noncoding RNA SERTAD2-3 inhibits osteosarcoma proliferation and migration by competitively binding miR-29c. Genet Test Mol Biomarkers. 2020;24(2):67–72. doi:10.1089/gtmb.2019.0164
30. Chen C, Li H, Wang X, Wang L, Zeng Q. Lnc-LFAR1 affects intrahepatic cholangiocarcinoma proliferation, invasion, and EMT by regulating the TGFβ/Smad signaling pathway. Int J Clin Exp Pathol. 2019;12(7):2455–2461.
31. Huang Z, Zhao X, Wu X, et al. LncRNA UCA1 facilitated cell growth and invasion through the miR-206/CLOCK axis in glioma. Cancer Cell Int. 2019;19:316. doi:10.1186/s12935-019-1023-7
32. Tritto V, Ferrari L, Esposito S, et al. Non-coding RNA and tumor development in neurofibromatosis type 1: ANRIL Rs2151280 is associated with optic glioma development and a mild phenotype in neurofibromatosis type 1 patients. Genes. 2019;10(11):892. doi:10.3390/genes10110892
33. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034
34. Dang Y, Wei X, Xue L, Wen F, Gu J, Zheng H. Long non-coding RNA in glioma: target miRNA and signaling pathways. Clin Lab. 2018;64(6):887–894. doi:10.7754/Clin.Lab.2018.180107
35. Mesci A, Huang X, Taeb S, et al. Targeting of CCBE1 by miR-330-3p in human breast cancer promotes metastasis. Br J Cancer. 2017;116(10):1350–1357. doi:10.1038/bjc.2017.105
36. Guan A, Wang H, Li X, et al. MiR-330-3p inhibits gastric cancer progression through targeting MSI1. Am J Transl Res. 2016;8(11):4802–4811.
37. Jin Z, Jia B, Tan L, Liu Y. miR-330-3p suppresses liver cancer cell migration by targeting MAP2K1. Oncol Lett. 2019;18(1):314–320. doi:10.3892/ol.2019.10280
38. Glenfield C, McLysaght A. Pseudogenes provide evolutionary evidence for the competitive endogenous RNA hypothesis. Mol Biol Evol. 2018;35(12):2886–2899. doi:10.1093/molbev/msy183
39. Xiong Z, Wang L, Wang Q, Yuan Y. LncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J Cell Mol Med. 2018;22(8):3929–3940. doi:10.1111/jcmm.13667
40. Zhang Y, Li Y, Wang J, Lei P. Long non-coding RNA ferritin heavy polypeptide 1 pseudogene 3 controls glioma cell proliferation and apoptosis via regulation of the microRNA-224-5p/tumor protein D52 axis. Mol Med Rep. 2018;18(5):4239–4246. doi:10.3892/mmr.2018.9491
41. Kudinov AE, Karanicolas J, Golemis EA, Boumber Y. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin Cancer Res. 2017;23(9):2143–2153. doi:10.1158/1078-0432.CCR-16-2728
42. Chen HY, Lin LT, Wang ML, et al. Musashi-1 promotes chemoresistant granule formation by PKR/eIF2α signalling cascade in refractory glioblastoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864(5Pt A):1850–1861. doi:10.1016/j.bbadis.2018.02.017
43. Chen HY, Lin LT, Wang ML, et al. Musashi-1 enhances glioblastoma cell migration and cytoskeletal dynamics through translational inhibition of Tensin3. Sci Rep. 2017;7(1):8710. doi:10.1038/s41598-017-09504-7
44. de Araujo PR, Gorthi A, da Silva AE, et al. Musashi1 impacts radio-resistance in glioblastoma by controlling DNA-protein kinase catalytic subunit. Am J Pathol. 2016;186(9):2271–2278. doi:10.1016/j.ajpath.2016.05.020
45. Chen HY, Lin LT, Wang ML, et al. Musashi-1 regulates AKT-derived IL-6 autocrinal/paracrinal malignancy and chemoresistance in glioblastoma. Oncotarget. 2016;7(27):42485–42501. doi:10.18632/oncotarget.9890
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.