Back to Journals » OncoTargets and Therapy » Volume 13
LINC01089 Blocks the Proliferation and Metastasis of Colorectal Cancer Cells via Regulating miR-27b-3p/HOXA10 Axis
Received 30 March 2020
Accepted for publication 29 July 2020
Published 19 August 2020 Volume 2020:13 Pages 8251—8260
DOI https://doi.org/10.2147/OTT.S256148
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Sanjeev K. Srivastava
Ming Li,1 Xufeng Guo2
1Department of Gastroenterology, Renmin Hospital of Wuhan University, Wuhan 430000, Hubei, People’s Republic of China; 2Cancer Center, Renmin Hospital of Wuhan University, Wuhan 430000, Hubei, People’s Republic of China
Correspondence: Xufeng Guo
Cancer Center, Renmin Hospital of Wuhan University, Wuhan 430000, Hubei, People’s Republic of China
Email [email protected]
Background: An increasing number of studies demonstrate that long non-coding RNAs (lncRNAs) are regulators in cancer biology. Nevertheless, the expression and mechanism of LINC01089 in colorectal cancer (CRC) remain unclear.
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was taken to investigate the expression levels of LINC01089 and miR-27b-3p in CRC tissues and cells. MTT method and transwell test were employed to assess the proliferation and invasion of CRC cells, respectively. Dual-luciferase activity reporter assay, RNA immunoprecipitation assay, Pearson’s correlation analysis, and Western blot were performed to investigate the regulatory mechanism of LINC01089/miR-27b-3p/HOXA10 axis in CRC.
Results: LINC01089 was down-regulated in CRC tissues and cell lines. LINC01089 overexpression impeded the proliferation and invasion of SW620 and LoVo cells, whereas LINC01089 knockdown increased the malignancy of SW480 and HT29 cells. Moreover, LINC01089 directly interacted with miR-27b-3p to repressed its expression and indirectly promoted the expression of HOXA10.
Conclusion: LINC01089 impedes the proliferation and invasion of colorectal cancer cells by adsorbing miR-27b-3p and up-regulating the expression of HOXA10.
Keywords: LINC01089, CRC, miR-27b-3p, HOXA10
Introduction
Colorectal cancer (CRC), the fourth major cause of cancer-related death around the world, has an annual incidence of approximately 1.4 million, and causes approximately 700,000 deaths each year.1 Surgery is usually utilized to treat CRC in early stage. For advanced patients with metastasis, chemotherapy and target therapy are the main treatments. As many as 40% of patients will recur after surgery, and the prognosis of these patients is poor.2
The majority of the human genomes are transcribed into non-coding RNAs (ncRNAs), like small ncRNAs and long ncRNAs (lncRNAs).3,4 LncRNAs are non-coding transcripts of >200 nucleotides without the ability of coding protein, and they regulate biological behaviors of cells through the interaction with DNA, RNA, and proteins.5,6 In tumorigenesis and cancer progression, lncRNAs play prominent roles.7–12 For example, lncRNA DILC is down-regulated in CRC tissues, and its high expression impedes cancer cell proliferation and metastasis, and it is a favorable indicator for the prognosis of CRC patients;9,10 the high expression of TTN-AS1 increases the malignancy of CRC cells and is associated with adverse clinicopathological features of CRC patients;11 the expression of lncRNA TCF7 in CRC is markedly linked to tumor growth, differentiation, and lymph node metastasis.12 However, the dysregulated lncRNAs in CRC have not been fully uncovered.
MicroRNAs (miRNAs) are small non-coding RNAs whose length are about 21–25nt. They directly bind to the 3ʹ-untranslated region (3ʹ-UTR) of target mRNAs to regulate gene expression.13 miRNAs exert an indispensable impact on a series of cell life activities, like cell cycle, differentiation, and proliferation. They can regulate many pathways associated with human diseases.14,15 A variety of miRNAs is abnormally expressed in CRC, and they regulate the malignant biological behaviors of cancer cells.16–20 For example, miR-27b-3p promotes migration and invasion of CRC cells by targeting homeobox A10 (HOXA10).20
The object of this research was to evaluate the expression of LINC01089, a rarely investigated lncRNA, in CRC tissues and cell lines, and to explore the function and mechanism of LINC01089 in CRC tumorigenesis. It was demonstrated that, LINC01089 was significantly down-regulated in CRC, and it suppressed the malignant biological behaviors of CRC cells via regulating miR-27b-3p/HOXA10 axis.
Materials and Methods
Clinical Data and Ethics Statement
CRC samples and adjacent normal tissues were harvested from 57 patients in Renmin Hospital of Wuhan University from 2015 April to 2018 April. All of the patients provided written informed consent for the collection and utilization of their tissue samples. All tissues were immediately frozen in liquid nitrogen at −196 °C after removal during surgery. The collection and use of the human samples in this study obtained the approval from the Research Ethics Committee of Renmin Hospital of Wuhan University.
Cell Line Culture
Human CRC cell lines (SW480, HT29, SW620, HCT116, as well as LoVo cells) and normal human colon epithelial cells (NCM460 cells) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Then Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA) was used to culture the cells. The medium contained 10% heat-inactivated fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Hyclone, Logan, UT, USA). These cells were placed in an incubator at 37°C with 5% CO2. The medium was changed every 3–4 d. Subculture was performed with 0.25% trypsin (Hyclone, Hyclone, Logan, UT, USA).
Cell Transfection
pcDNA empty vector (Vector), pcDNA-LINC01089 (LINC01089), siRNA normal control (si-NC), siRNAs against LINC01089 (si-LINC01089), miR-27b-3p mimic, and miR-27b-3p inhibitors were provided by GenePharma Co., Ltd. (Shanghai, China). SW480, HT29, SW620 and LoVo cells were harvested, re-suspended with serum-free medium and inoculated into a 6-well cell culture plate (2×105/well). After 24h of culture, the CRC cells were transfected with Lipofectamine ® 2000 (Invitrogen, Carlsbad, CA, USA) according to the supplier’s instructions. 12 h later, complete medium was added to continue the culture. After 24h, quantitative real-time polymerase chain reaction (qRT-PCR) was utilized to investigate the transfection efficiency.
qRT-PCR
Total RNA from tissues and cells was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). SuperScript First Strand cDNA System (Invitrogen, Carlsbad, CA, USA) was utilized to reversely transcribe 1 μg of total RNA into complementary DNA (cDNA). With cDNA as template, qRT-PCR was performed on a ABI 7500 Fast Real-Time PCR System with SYBR®PremixExTaqTM kit (Takara, Dalian, China). The primers used in this work were as follows: LINC01089: 5′-GCAGTAAACAGTCCTCAGCGAAG-3′(forward) and 5′-CGGTGCCATGGAGTCTAGAAGAT-3′(reverse); HOXA10, 5′-AGATATTGTCCTAAGTGTCAAGTCCTGA-3′(forward) and 5′-GCCATTTCGAGCAGTGGG-3′(reverse); U6, 5′-CTCGCTTCGCRCAGCACA-3′(forward) and 5′-AACGCTTCACGAATTTGCGT-3′(reverse); GAPDH: 5′-TGTCCGTCGTGGATCTGA-3′(forward) and 5′-TTGCTGTTGAAGTCGCAGGAGG-3′(reverse); hsa-miR-27b-3p: 5ʹ-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCAGAACT-3′(forward) and 5′-ACACTCCAGCTGGGTTCACAGTGGCTAAG-3′(reverse). The expression levels of LINC01089, miR-27b-3p, and HOXA10 were measured employing the 2− ΔΔCT method.
3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT) Experiment
CRC cells were inoculated in a 96-well plate (2×103 cells/well), and cultured for different times (1 d, 2 d, 3 d, 4 d, respectively). At each time point, 20 μL MTT solution (5 mg/mL, Beyotime, Shanghai, China) was added into the wells. After that, the cells were incubated with the MTT solution for 4 h. Subsequently, after the medium was discarded, 150 μL dimethyl sulfoxide was added into each well. The plate was shaken for 10 min to resolve the MTT formazan crystals. Next, a scanning multi-well spectrophotometer was utilized to measure the optical density (OD) value in each well at a wavelength of 490 nm.
Transwell Experiment
After CRC cells were dispersed with 0.25% trypsin, the CRC cells were centrifuged, re-suspended with serum-free medium, and inoculated into the upper transwell chamber (8 μm pore size, BD Biosciences, CA, USA) placed in 24-well plates, which contained complete medium (600 μ/L per well). The cells were cultured in 37 °C for 24 h. After that, the cells on the upper surface of membrane were gently wiped off with a cotton swab, and 4% paraformaldehyde was employed to fix the invaded cells. Furthermore, the cells were stained with 0.5% crystal violet solution. After rinsed with tap water and dried, the cells were observed under an inverted microscope and counted.
Western Blot
Cells were immersed in pre-cooled RIPA lysis buffer (Solarbio, Beijing, China), and placed on ice for 30 min. After centrifugation, the supernatant was collected. The samples were added with loading buffer, and denatured in boiling water. After the separation of protein samples by SDS-PAGE, the proteins were transferred to the polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA). Next, the membrane was incubated in blocking buffer (5% skimmed milk) for 1 h at room temperature, and subsequently washed with TBST solution. Next, the membrane was incubated overnight with primary antibodies at 4 °C. The primary antibodies included: anti-HOXA10 (Abcam, Cambridge, ab191470, 1:500), anti-GAPDH (Abcam, Cambridge, ab8245, 1:3000). After the PVDF membrane was rinsed with TBST solution again, it was incubated with horseradish peroxidase conjugated secondary antibody (Abcam, ab150117, 1:2000) at room temperature for 1 h. Finally, hypersensitive ECL kit (Beyotime, Shanghai, China) was utilized for developing protein bands on an X-ray film.
Dual-Luciferase Reporter Gene Assay
Luciferase reporter assay was carried out by with dual-luciferase reporter assay system (Promega, Madison, WI, USA). The target fragments of wild type LINC01089/3ʹUTR of HOXA10 and mutant LINC01089/3ʹUTR of HOXA10 were synthesized and integrated into psi-CHECK2 reporter vector (Promega, Madison, WI, USA) to construct the recombinant luciferase reporter vectors. The recombinant reporter vectors were co-transfected with 293T cells with miR-27b-3p mimics or control microRNAs. Forty-eight hours after transfection, luciferase activity was determined according to the manufacturer’s instructions.
RNA Immunoprecipitation (RIP) Assay
EZ-Magna RNA binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) was employed to perform RIP experiment. In brief, cells were harvested and resuspended in RIP lysis buffer. The cell extracts were subsequently incubated with RIP buffer which contained magnetic beads conjugated with human anti-Ago2 antibody or mouse IgG overnight. After the magnetic beads were washed 3 times, they were incubated with proteinase K. Subsequently, total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Finally, the relative enrichment of LINC01089 and miR-27b-3p were determined by qRT-PCR analysis.
Statistical Analysis
SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA) was utilized for data analysis. All of the experiments were conducted independently at least 3 times, and the measurement data were shown as mean ± standard deviation (x ± s). Multivariate comparisons were carried out by one-way analysis of variance. Additionally, comparisons between two groups were carried out employing student’s t-tests. The difference had statistical significance at P <0.05.
Result
LINC01089 Was Down-Regulated in CRC Tissues and Cell Lines
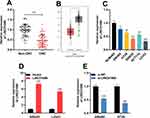
Firstly, qRT-PCR results illustrated that the expression of LINC01089 in CRC tissues was markedly lower than that in adjacent colonic tissues (Figure 1A). Next, GEPIA database (http://gepia.cancer-pku.cn/) was employed to analyze the expression of LINC01089 in CRC tissues, and it was demonstrated that LINC01089 was differentially expressed in CRC cancer tissues and normal colonic tissues, and the expression of LINC01089 in cancer tissues was markedly lower than normal tissues (Figure 1B). Additionally, the expressions of LINC01089 in CRC cell lines (SW480, HT29, SW620, HCT116, as well as LoVo cells) were also markedly lower than in immortalized colonic epithelial cells (NCM460 cells) (Figure 1C).
LINC01089 Impeded the Proliferation and Invasion of CRC Cells
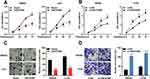
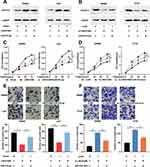
According to the above results, among CRC cells, the expression of LINC01089 was comparatively lower in SW620 and LoVo cells, whereas it was higher in SW480 and HT29 cells. Therefore, LINC01089 was overexpressed in SW620 and LoVo cells with LINC01089 overexpression plasmid, and SW480 and HT29 cells were transfected with LINC01089 siRNA to establish LINC01089 knockdown models (Figure 1D and E). MTT method and transwell method were taken to investigate the proliferation and invasion of CRC cells, respectively. As shown, LINC01089 overexpression significantly impeded the proliferation of SW620 and LoVo cells (Figure 2A), whereas LINC01089 knockdown markedly promoted the proliferation of SW480 and HT29 cells (Figure 2B). Moreover, SW620 and LoVo cells with LINC01089 overexpression were less invasive than the control group (Figure 2C), whereas SW480 and HT29 cells with LINC01089 knockdown were more invasive than the control group (Figure 2D).
LINC01089 Directly Acted on miR-27b-3p in CRC Cells
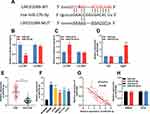
Next, StarBase (http://starbase.sysu.edu.cn/) was utilized to perform a bioinformatic analysis, and it predicted that LINC01089 contained a binding sequence for miR-27b-3p (Figure 3A, Supplementary Material). To validate this prediction, dual-luciferase reporter assay was performed, and it showed that the luciferase activity of wild-type LINC01089 reporter vector was reduced by miR-27b-3p, whereas the luciferase activity of mutant LINC01089 reporter vector was not markedly changed by miR-27b-3p; while miR-27b-3p inhibitors promoted the luciferase activity of wile-type LINC01089 reporter vector, and did not change the luciferase activity of mutant LINC01089 reporter vector (Figure 3B and C). Subsequently, RIP assay confirmed that, compared with IgG, LINC01089 and miR-27b-3p were enriched in Ago2-containing microribonucleoproteins enriched by anti-Ago2 antibodies (Figure 3D). Moreover, qRT-PCR was utilized to investigate the expression of miR-27b-3p in CRC tissues and cell lines. It was demonstrated that compared with normal tissues and NCM460 cells, the expression of miR-27b-3p markedly increased in CRC cancer tissues (Figure 3E) and cell lines (Figure 3F). What’s more, Pearson’s correlation analysis indicated a strong negative correlation between the LINC01089 expression and miR-27b-3p expression in CRC tissue samples (R2 = 0.6714, P<0.001) (Figure 3G). These results suggested that LINC01089 could function as a molecular sponge of miR-27b-3p to repress it. Additionally, it was demonstrated that the expression of LINC01089 in CRC cells was not changed by the transfection of miR-27b-3p mimics or inhibitors (Figure 3H), which implied that the regulatory relationship between LINC01089 and miR-27b-3p was unidirectional.
HOXA10 Was a Target Gene of miR-27b-3p in CRC
Next, bioinformatic analysis was performed to predict targets of miR-27b-3p with TargetScan (http://www.targetscan.org/vert_72/), and it showed that HOXA10 was a possible downstream target of miR-27b-3p (Figure 4A). Dual-luciferase reporter assay showed that miR-27b-3p inhibited the luciferase activity of wild-type HOXA10 reporter vector, and miR-27b-3p did not change the luciferase activity of mutant HOXA10 reporter vector; while miR-27b-3p inhibitors promoted the luciferase activity of wile-type HOXA10 reporter vector, and did not change the luciferase activity of mutant HOXA10 reporter vector (Figure 4B and C). These data validated the targeting relationship between miR-27b-3p and the 3ʹUTR of HOXA10. These results were consistent with the demonstrations in the previous report.20 Subsequently, qRT-PCR was used to detect the expression of HOXA10A mRNA in CRC tissues, and the results suggested that compared with that in adjacent tissues, it was markedly down-regulated in cancerous tissues (Figure 4D), and Pearson’s correlation analysis confirmed that expression of HOXA10 was positively correlated with expression of LINC01089 in CRC tissues (R2 = 0.7445, P<0.001) (Figure 4E). Additionally, Western blot suggested that the transfection of miR-27-3b mimics inhibited the expression of HOXA10 in CRC cells, while miR-27b-3p inhibitors promoted the expression of HOXA10 in CRC cells (Figure 4F). These results suggested that HOXA10 was negatively regulated by miR-27b-3p, and LINC01089 probably positively regulated the expression of HOXA10 expression in CRC via repressing miR-27b-3p.
LINC01089 Regulated miR-27b-3p/HOXA10 Axis to Suppress the Proliferation and Invasion of CRC Cells
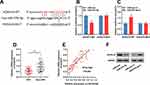
To study whether LINC01089 affected the CRC progression by modulating the miR-27b-3p/HOXA10 axis, rescue experiments were performed. Western blot showed that protein expression of HOXA10 was up-regulated in SW620 and LoVo cells after LINC01089 overexpression, and the expression of HOXA10 was partially reduced after the co-transfection of miR-27b-3p mimics (Figure 5A); the expression of HOXA10 protein in SW480 and HT29 cells were decreased after LINC01089 was knocked down, and the expression of HOXA10 partially recovered after co-transfection with miR-27b-3p inhibitors (Figure 5B). Additionally, functional experiments showed that LINC01089 overexpression markedly impeded the proliferation and invasion of SW620 and LoVo cells, and miR-27b-3p mimics partially reversed the inhibitory impacts on the proliferation and invasion of CRC cells induced by LINC01089; on the other hand, LINC01089 knockdown markedly facilitated the proliferation and invasion of SW480 and HT29 cells, whereas miR-27b-3p inhibitors partially attenuated the promotion of proliferation and invasion induced by LINC01089 knockdown (Figure 5C–F). These data proved that LINC01089 could impede the proliferation and invasion of CRC cells by regulating miR-27b-3p/HOXA10 axis.
Discussion
CRC, one of the most common malignancies of the digestive tract, is driven by a series of genetic changes, and it seriously endangers human health.21,22 As an important regulator related to tumorigenesis and cancer progression, the function of lncRNA in CRC cells has not been totally elucidated.
LINC01089 (also known as LncRNA Inhibiting Metastasis, LIMT) was firstly reported to be suppressed by epidermal growth factor in breast cancer, and its dysregulation was linked to cancer progression.23 Recent studies have shown that LINC01089 exerts an inhibitory effect on some cancers. In breast cancer, LINC01089 overexpression impedes cancer metastasis via regulating Wnt/β-catenin signaling.24 Moreover, LINC01089 is down-regulated in glioma tissues, and its down-regulation predicts unfavorable prognosis of the patients; additionally, and its restoration impedes the malignant biological behaviors of glioma cells.25 Nevertheless, previously, the function and underlying mechanism of LINC01089 during CRC progression had not been clarified. For the first time, this study confirmed that LINC01089 was under-expressed in CRC tissues and cells. It was also demonstrated that LINC01089 suppressed the malignant biological behaviors of CRC cells. These data indicated that LINC01089 played a tumor suppressive role in CRC, which is similar with its function in other cancers.23–25
LncRNA can share miRNA response elements of mRNA and competitively bind with miRNAs to reduce its availability, functioning as competitive endogenous RNA (ceRNA), thereby exerting an impact on the translation of mRNAs.26,27 The functions of miR-27b-3p in cancer biology have been reported previously, and its role in different cancers is specific. In breast cancer, lung cancer, gastric cancer and endometrial cancer, it inhibits cancer cell proliferation and metastasis;28–31 however, in anaplastic thyroid cancer, the up-regulation of miR-27b-3p contributes to the drug resistance of cancer cells to doxorubicin,32 and in CRC, miR-27b-3p functions as an oncomiR to promote migration and invasion by targeting HOXA10.20 In this study, it was hypothesized that LINC01089 could probably participate in CRC progression as a ceRNA. Bioinformatic analysis, dual-luciferase reporter assay and RIP assay confirmed that LINC01089 could sponge miR-27b-3p to repress its expression. It was also observed that the expression of miR-27b-3p and LINC01089 was negatively correlated in CRC. Importantly, functional experiments showed that miR-27b-3p could reverse the function of LINC01089 in regulating the malignant phenotypes of CRC cells. These results suggested that the down-regulation of LINC01089 in CRC contributed to the dysregulation of miR-27b-3p, and the tumor-suppressive function of LINC01089 in CRC was at least partially dependent on miR-27b-3p.
HOXA10 is a key transcription factor in regulating embryonic development, and it also functions in fertility, embryo viability, and regulation of hematopoietic lineage commitment.33 In recent years, the role of HOXA10 in cancer biology is unveiled gradually. In the tumorigenesis of myeloid leukemia, it is associated with self-renewal capability of myeloid progenitors.34 HOXA10 overexpression in ovarian cancer promotes the epithelial-mesenchymal transition of cancer cells.35,36 In gastric cancer, expression of HOXA10 is up-regulated and it exerts oncogenic functions by activating the JAK1/STAT3 signaling pathway.37 Moreover, the defect in HOXA10 impedes the proliferation of testicular cancer cells by blocking cell cycle process.38 However, in breast cancer, HOXA10 is reported to be tumor-suppressive, and it reduces the invasiveness of breast cancer cells by up-regulating p53.39 In CRC, HOXA10 inhibits cancer progression, and it has been reported to be negatively regulated by oncomiRs including miR-27b-3p and miR-544a.20,40 This present research verified that HOXA10 was a downstream target of miR-27b-3p, and this conclusion is consistent with previous report.20 Further experiments verified that LINC01089 had a positive regulatory effect on the expression of HOXA10 by targeting miR-27b-3p. These data indicated that HOXA10 participated in blocking CRC progression mediated by LINC01089.
In summary, this study demonstrates that LINC01089 is down-regulated in CRC, working as a molecular sponge for miR-27b-3p to modulate the expression of HOXA10, and has an inhibitory effect on the proliferation and invasion of CRC cells. Therefore, LINC01089 can be a promising target for the diagnosis and treatment of CRC.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
The authors declare that they have no competing interests.
References
1. Moreno EC, Pascual A, Prieto-Cuadra D, et al. Novel molecular characterization of colorectal primary T umors based on miRNAs. Cancers. 2019;11:3. doi:10.3390/cancers11030346
2. Cantero-Cid R, Casas-Martin J, Hernández-Jiménez E, et al. PD-L1/PD-1 crosstalk in colorectal cancer: are we targeting the right cells? BMC Cancer. 2018;18(1):945. doi:10.1186/s12885-018-4853-0
3. Yuan Y, Sun S, Jiao N, Shu Y, Zhang Y. Upregulation of HOXA10 protein expression predicts poor prognosis for colorectal cancer. Genet Test Mol Biomarkers. 2018;22(6):390–397. doi:10.1089/gtmb.2017.0240
4. Yang L, Xie N, Huang J, et al. SIK1-LNC represses the proliferative, migrative, and invasive abilities of lung cancer cells. Onco Targets Ther. 2018;11:4197–4206. doi:10.2147/OTT.S165278
5. Wang YL, Liu JY, Yang JE, et al. Lnc-UCID promotes G1/S transition and hepatoma growth by preventing DHX9-mediated CDK6 down-regulation. Hepatology. 2019;70(1):259–275. doi:10.1002/hep.30613
6. Yoshimura H, Matsuda Y, Yamamoto M, Kamiya S, Ishiwata T. Expression and role of long non-coding RNA H19 in carcinogenesis. Front Biosci. 2018;23:614–625. doi:10.2741/4608
7. Cho SW, Xu J, Sun R, et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell. 2018;173(6):1398–1412. doi:10.1016/j.cell.2018.03.068
8. Ji D, Zhong X, Jiang X, et al. The role of long non-coding RNA AFAP1-AS1 in human malignant tumors. Pathol Res Pract. 2018;214(10):1524–1531. doi:10.1016/j.prp.2018.08.014
9. Gu LQ, Xing XL, Cai H, et al. Long non-coding RNA DILC suppresses cell proliferation and metastasis in colorectal cancer. Gene. 2018;666:18–26. doi:10.1016/j.gene.2018.03.100
10. Li QG, Xu XQ, Zhou DY, et al. Long non-coding RNA DILC as a potentially useful biomarker for the diagnosis and prognosis of colorectal cancer. Eur Rev Med Pharmacol Sci. 2019;23(8):3320–3325. doi:10.26355/eurrev_201904_17694
11. Cui ZH, Han BB, Wang XR, Li ZW, Wang JX, Lv YF. Long non-coding RNA TTN-AS1 promotes the proliferation and invasion of colorectal cancer cells by activating miR-497-mediated PI3K/Akt/mTOR signaling. Onco Targets Ther. 2019;12:11531–11539. doi:10.2147/OTT.S229104
12. Jin FS, Wang HM, Song XY. Long non-coding RNA TCF7 predicts the progression and facilitates the growth and metastasis of colorectal cancer. Mol Med Rep. 2018;17(5):6902–6908. doi:10.3892/mmr.2018.8708
13. Wang S, Zhang G, Zheng W, et al. MiR-454-3p and miR-374b-5p suppress migration and invasion of bladder cancer cells through targetting ZEB2. Biosci Rep. 2018;38(6). doi:10.1042/BSR20181436
14. Li T, Pan H, Li R. The dual regulatory role of miR-204 in cancer. Tumour Biol. 2016;37(9):11667–11677. doi:10.1007/s13277-016-5144-5
15. Mohammadi Torbati P, Asadi F, Fard-Esfahani P. Circulating miR-20a and miR-26a as biomarkers in prostate cancer. Asian Pac J Cancer Prev. 2019;20(5):1453–1456. doi:10.31557/APJCP.2019.20.5.1453
16. Chen E, Li Q, Wang H, et al. MiR-32 promotes tumorigenesis of colorectal cancer by targeting BMP5. Biomed Pharmacother. 2018;106:1046–1051. doi:10.1016/j.biopha.2018.07.050
17. Chen E, Li Q, Wang H, Yang F, Min L, Yang J. MiR-92a promotes tumorigenesis of colorectal cancer, a transcriptomic and functional based study. Biomed Pharmacother. 2018;106:1370–1377. doi:10.1016/j.biopha.2018.07.098
18. Li Y, Duo Y, Bi J, et al. Targeted delivery of anti-miR-155 by functionalized mesoporous silica nanoparticles for colorectal cancer therapy. Int J Nanomedicine. 2018;13:1241–1256. doi:10.2147/IJN.S158290
19. O’Brien SJ, Carter JV, Burton JF, et al. The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: a systematic review. Int J Cancer. 2018;142(12):2501–2511. doi:10.1002/ijc.31282
20. Yang X, Chen J, Liao Y, et al. MiR-27b-3p promotes migration and invasion in colorectal cancer cells by targeting HOXA10. Biosci Rep. 2019;39(12). doi:10.1042/BSR20191087
21. Li B, Shi C, Zhao J, Li B. Long noncoding RNA CCAT1 functions as a ceRNA to antagonize the effect of miR-410 on the down-regulation of ITPKB in human HCT-116 and HCT-8 cells. Oncotarget. 2017;8(54):92855–92863. doi:10.18632/oncotarget.21612
22. Ruiz-López L, Blancas I, Garrido JM, et al. The role of exosomes on colorectal cancer: a review. J Gastroenterol Hepatol. 2018;33(4):792–799. doi:10.1111/jgh.14049
23. Sas-Chen A, Aure MR, Leibovich L, et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol Med. 2016;8(9):1052–1064. doi:10.15252/emmm.201606198
24. Yuan H, Qin Y, Zeng B, et al. Long noncoding RNA LINC01089 predicts clinical prognosis and inhibits cell proliferation and invasion through the Wnt/β-catenin signaling pathway in breast cancer. Onco Targets Ther. 2019;12:4883–4895. doi:10.2147/OTT.S208830
25. Gu J, Xu F, Dang Y, Bu X. Long non-coding RNA 001089 is a prognostic marker and inhibits glioma cells proliferation and invasion. Clin Lab. 2019;65(3). doi:10.7754/Clin.Lab.2018.180817
26. Shuwen H, Qing Z, Yan Z, Xi Y. Competitive endogenous RNA in colorectal cancer: a systematic review. Gene. 2018;645:157–162. doi:10.1016/j.gene.2017.12.036
27. Wang H, Wang G, Gao Y, et al. Lnc-SNHG1 activates the TGFBR2/SMAD3 and RAB11A/Wnt/β-catenin pathway by sponging MiR-302/372/373/520 in invasive pituitary tumors. Cell Physiol Biochem. 2018;48(3):1291–1303. doi:10.1159/000492089
28. Chen D, Si W, Shen J, et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018;9(2):188. doi:10.1038/s41419-017-0211-4
29. Sun W, Zhang LW, Yan RR, Yang Y, Meng XL. LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. Onco Targets Ther. 2019;12:3945–3954. doi:10.2147/OTT.S196865
30. Tao J, Zhi X, Zhang X, et al. miR-27b-3p suppresses cell proliferation through targeting receptor tyrosine kinase like orphan receptor 1 in gastric cancer. J Exp Clin Cancer Res. 2015;34(1):139. doi:10.1186/s13046-015-0253-3
31. Liu L, Hu J, Yu T, et al. miR-27b-3p/MARCH7 regulates invasion and metastasis of endometrial cancer cells through snail-mediated pathway. Acta Biochim Biophys Sin (Shanghai). 2019;51(5):492–500. doi:10.1093/abbs/gmz030
32. Xu Y, Han YF, Ye B, et al. miR-27b-3p is involved in doxorubicin resistance of human anaplastic thyroid cancer cells via targeting peroxisome proliferator-activated receptor gamma. Basic Clin Pharmacol Toxicol. 2018;123(6):670–677. doi:10.1111/bcpt.13076
33. Zanatta A, Rocha AM, Carvalho FM, et al. The role of the Hoxa10/HOXA10 gene in the etiology of endometriosis and its related infertility: a review. J Assist Reprod Genet. 2010;27(12):701–710. doi:10.1007/s10815-010-9471-y
34. Oakley K, Han Y, Vishwakarma BA, et al. Setbp1 promotes the self-renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10. Blood. 2012;119(25):6099–6108.33. doi:10.1182/blood-2011-10-388710
35. Liu J, Jiang Y, Wan Y, Zhou S, Thapa S, Cheng W. MicroRNA665 suppresses the growth and migration of ovarian cancer cells by targeting HOXA10. Mol Med Rep. 2018;18(3):2661–2668. doi:10.3892/mmr.2018.9252
36. Zhang HY, Li JH, Li G, Wang SR. Activation of ARK5/miR-1181/HOXA10 axis promotes epithelial-mesenchymal transition in ovarian cancer. Oncol Rep. 2015;34(3):1193–1202. doi:10.3892/or.2015.4113
37. Chen W, Wu G, Zhu Y, et al. HOXA10 deteriorates gastric cancer through activating JAK1/STAT3 signaling pathway. Cancer Manag Res. 2019;11:6625–6635. doi:10.2147/CMAR.S201342
38. Chen R, Li H, Li Y, et al. Loss of nuclear functions of HOXA10 is associated with testicular cancer proliferation. Front Oncol. 2018;8:594. doi:10.3389/fonc.2018.00594
39. Chu MC, Selam FB, Taylor HS. HOXA10 regulates p53 expression and matrigel invasion in human breast cancer cells. Cancer Biol Ther. 2004;3(6):568–572. doi:10.4161/cbt.3.6.848
40. Sun S, Su C, Zhu Y, et al. MicroRNA-544a regulates migration and invasion in colorectal cancer cells via regulation of homeobox A10. Dig Dis Sci. 2016;61(9):2535–2544. doi:10.1007/s10620-016-4186-2
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.