Back to Journals » OncoTargets and Therapy » Volume 13
LINC01087 is Highly Expressed in Breast Cancer and Regulates the Malignant Behavior of Cancer Cells Through miR-335-5p/Rock1
Authors She JK, Fu DN, Zhen D, Gong GH, Zhang B
Received 29 March 2020
Accepted for publication 14 August 2020
Published 30 September 2020 Volume 2020:13 Pages 9771—9783
DOI https://doi.org/10.2147/OTT.S255994
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Ji-Kai She,1,2 Dan-Ni Fu,1,2 Dong Zhen,1,2 Guo-Hua Gong,2,3 Bin Zhang2,3
1Medicinal Chemistry and Pharmacology Institute, Inner Mongolia University for the Nationalities, Tongliao, Inner Mongolia, People’s Republic of China; 2Inner Mongolia Key Laboratory of Mongolian Medicine Pharmacology for Cardio-Cerebral Vascular System, Tongliao, Inner Mongolia, People’s Republic of China; 3First Clinical Medical of Inner Mongolia University for Nationalities, Tongliao, Inner Mongolia, People’s Republic of China
Correspondence: Bin Zhang; Guo-Hua Gong
Inner Mongolia Key Laboratory of Mongolian Medicine Pharmacology for Cardio-Cerebral Vascular System, Tongliao, Inner Mongolia, People’s Republic of China
Email [email protected]; [email protected]
Purpose: Long non-coding RNA is involved in the genesis and development of various tumors, and it has been found through database screening that LINC01087 is highly expressed in breast cancer (BC), but mechanisms of LINC01087 in BC are still under investigation. Therefore, this study aimed to study relevant mechanisms of LINC01087 in BC to provide potential therapeutic targets for the disease in clinic practice.
Patients and Methods: The qRT-PCR assay was applied to determine the LINC01087 expression in BC, and the cell counting kit-8 (CCK8) assay, transwell assay, and flow cytometry were used to analyze the proliferation, apoptosis, and invasion of breast cancer cells (BCCs), respectively. The Western blot assay was used to determine the ROCK1 expression, and the luciferase reporter gene assay, RNA-binding protein immunoprecipitation (RIP), and RNA pull-down assays were applied to study the interaction between LINC01087 and miR-335-5p. Moreover, tumor xenotransplantation was conducted in nude mice to explore the effects of LINC01087 on BCCs.
Results: The qRT-PCR assay revealed that the LINC01087 expression in BC tissues was higher than that in corresponding tumor-adjacent tissues, and survival analysis revealed an unfavorable prognosis of patients with high expression of LINC01087. Down-regulation of LINC01087 could slow down the proliferation, invasion, and migration of BCCs and accelerate apoptosis of them in vitro. Luciferase reporter gene assay results revealed that LINC01087 enhanced the expression of ROCK1 by regulating miR-335-5p, and LINC01087 could be adopted as a miR-335-5p sponge to inhibit ROCK1 expression.
Conclusion: LINC01087 is overexpressed in cases with BC, and patients with high expression of it suffer a poor survival. Furthermore, LINC01087 can act as a miR-335-5p sponge to affect the expression of ROCK1 and affect the invasion and migration of BCCs.
Keywords: migration, LINC01087, miR-335-5p, ROCK1, breast cancer, ceRNA
Core Tip
In our study, it was confirmed that LINC01087 was highly expressed in breast cancer according to the database and clinical verification, and the 5-year survival of patients with high expression of it was poor. We also found that LINC01087 could act as a miR-335-5p sponge to regulate the ROCK1 expression and the invasion and migration of breast cancer cells.
Introduction
Breast cancer (BC), a malignant tumor with a high incidence among women, is considered to be the major cause of cancer-related death among women in the world.1,2 According to the latest tumor epidemiology statistics,3 there were more than 2,000,000 new BC patients (11.6%) and about 600,000 patients dying of it in 2018 (6.6%). At present, the screening for BC is wildly applied, so more and more early BC has been found, but the mortality of the cancer is still terribly high.4,5 In recent years, with the continuous improvement of medical level and treatment methods for BC, most patients can be treated by surgery. However, some patients can only be treated through radiotherapy and chemotherapy to improve prognosis, because they have already missed the optimal treatment timing, and many BC patients have suffered failed chemoradiotherapy, because they have no significant response to initial chemoradiotherapy.6–8 Therefore, it is urgent to investigate new mechanisms of BC.
Non-coding RNA (ncRNA) is a crucial part of the development and progression of tumors, which is involved in the biological functions of tumors, such as proliferation, invasion, and migration.9,10 Currently, the most widely studied ncRNA is microRNA (miR). Studies have found that miR is involved in tumor pathways and acts as a regulatory factor in tumor development and progression.11,12 With the deepening of research, long non-coding RNA (LncRNA) has been found to be important in the pathogenesis of various cancers including lung cancer, liver cancer, and BC.13–15 LncRNA is longer than 200nt. Studies have revealed that lncRNA affects gene expression at transcription and post-transcription levels, and it accelerates tumor growth by regulating the interaction among DNA, protein, and RNA and driving the biological function changes of various tumor cells.16,17 LINC01087 is a member of the lncRNA family, but there are few studies on it. We analyzed the common potential differential lncRNA expression based on the Cancer Genome Atlas (TCGA) and the gene expression omnibus (GEO) database, finding that LINC01087 was highly expressed in cases with BC, and LINC01087 was expected to be a potential target for BC.
Therefore, this study explored the value and related mechanisms of LINC01087 in BC to find new potential targets for the clinical treatment of BC.
Methods and Materials
Download and Analysis of Database Data
Corresponding data about lncRNA expression in BC were downloaded from https://portal.gdc.cancer.gov/and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113851. Data downloaded from TCGA were integrated to generate matrix file mRNA.symbol.txt, and converted through Log (X+1, 2). The edgR package was adopted to analyze data differences, and corresponding thermographies and volcano plots were drawn. Data downloaded from the GEO were first compared with Gene by blast, and then integrated into matrix file biotype.txt. The limma package was applied to analyze data differences, and corresponding thermographies and volcano plots were also drawn.
Collection of Clinical Samples
A total of 58 BC patients treated in Inner Mongolia Key Laboratory of Mongolian Medicine Pharmacology for Cardio-Cerebral Vascular System from May 2013 to May 2014 were enrolled, and their tumor tissues and corresponding tumor-adjacent tissues were sampled, transported by liquid nitrogen, and stored at −80°C. The inclusion criteria of the patients were as follows: Patients diagnosed with BC based on pathology and imaging examination, and patients meeting the version 7 TNM staging criteria released by the American Joint Committee on Cancer (AJCC) in 2009,18 patients who signed informed consent forms after understanding this study, and patients willing to cooperate with the treatment and follow-up. The exclusion criteria of them were as follows: Patients with other comorbid tumors, patients whose expected survival time were less than 3 months, patients who had received anti-tumor treatment before this treatment, and patients with triple-negative breast cancer (TNBC). The patients were followed up by telephone and based on electronic medical records of outpatient reexamination in January, March, June, September, and December each year. This study was carried out after relevant approval documents have been obtained from the Ethics Committee of Inner Mongolia Key Laboratory of Mongolian Medicine Pharmacology for Cardio-Cerebral Vascular System and was in accordance with the Helsinki Declaration.
Cell Culture
BCC lines (MDA-MB-231, MCF-7, T47D, and BT-549) and normal cell lines (MCF-10A) from the American Type Culture Collection (ATCC) were determined using short tandem repeat (STR), and their mycoplasma contamination was detected. The cells were incubated in dulbecco’s modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS) (Gibco, the United States) and 100 U/mL Penicillin/Streptomycin (Gibco, United State) in a thermostat with 5%CO2 at 37°C.
Cell Transfection
MDA-MB-231 and MCF-7 cell lines in the logarithmic growth phase were transfected after the cells lines were cultured in a 6-well plate at 1*106 per well for 24 h, and the cell confluency reached 90%. Si-LINC01087, miR-335-5p-mimics, miR-335-5p inhibit, si-ROCK1, and pcDNA-ROCK1 were designed and synthesized by Shanghai Sangon Biotech Co., Ltd. The vectors, miR mimics, and inhibitors were transfected into the cells with a Lipofectamine 2000 Kit (Invitrogen™, United States) in strict accordance with the kit instructions.
RNA Extraction and Real-Time qPCR
Total RNA was extracted from the tissues and cells sampled from the patients and tumor tissues sampled from mice using a TRIzol Kit (Invitrogen Company, United States), and its concentration, purity, and integrity were detected using an ultraviolet spectrophotometer and agarose gel electrophoresis. Subsequently, the RNA was reversely transcribed to cDNA with a TaqMan™ Reverse transcription Kit (Invitrogen Company, the United States) in strict accordance with the kit instructions, and the obtained cDNA was used for later analysis. PCR amplification was carried out with a PrimeScript RT Master Mix kit (Takarabo Company, Japan) in 20 μL total reaction volume containing 10 μL SYBR qPCR Mix, 0.8 μL upstream and downstream primers, respectively, 2 μL cDNA product, 0.4 μL 50×ROX reference dye, and RNase-free water added to adjust the volume, and PCR was carried out through pre-denaturation at 95°C for 60 s, followed by 40 cycles of denaturation at 95°C for 30 s, and annealing and extension at 60°C for 40 s. In the experiment, three parallel repeated wells were set, and each sample was detected repeatedly three times. Data in this study were analyzed using 2−ΔΔct with U6 and GADPH as internal references.19 The PCR instrument was a 7500PCR instrument from the ABI Company.
Dual-Luciferase Reporter Assay
Complementary DNA fragments containing wild-type (LINC01087-WT) or mutant-type LINC01087 (LINC01087-MUT) fragments were subcloned to the downstream of luciferase genes in psi-CHECK2 luciferase reporter vectors. Wild-type ROCK1 (ROCK1-WT) and corresponding mutant-type ROCK1 (ROCK1-MUT) fragments were established, and LINC01087/ROCK1 report vectors (Invitrogen, United States) and miR-335-5p mimics or miR-335-5p inhibitors were co-transfected into MDA-MB-231 cells using transfection reagents according to the above steps. After 48 hours of transfection, a dual-luciferase reporter assay kit (Promega, United States) was used to detect the firefly luciferase activity and renin-luciferase activity in cell lysates.
RNA-Binding Protein Immunoprecipitation (RIP)
RIP analysis was carried out through a Magna RNA-binding Protein Immunoprecipitation Kit (Millipore, United States), and RBE cells were lysed in RIP lysis buffer. Subsequently, 100 μL whole-cell extract was cultured with magnetic beads bound to human anti-Ago2 antibody or normal mouse immunoglobulin G (IgG) (Millipore, United States) at 4°C overnight, and the protein in the samples was digested with proteinase K through incubation, and finally, the immunoprecipitated RNA was separated by TRIzol Reagent and used for qRT-PCR analysis.
RNA Pull-Down Assay
The lnc LINC01087 probe was labeled with biotin using transcription, and the probe was incubated with MCF-7 cytoplasmic lysate using the transcription method to form lncRNA-microRNA complex coordination compound. The compound was bound to magnetic beads through chain affinity to separate it out from other components. Finally, the miR-335-5p family members pulled down were detected by compound elution and qRT-PCR.
Western Blot Assay
The protein in the collected cells was lysed with RIPA buffer (Cell Signal Technology, Inc., Massachusetts, United States). The protein concentration was determined with a Bicinchoninic Acid (BCA) Kit (Beyotime Biotechnology, Shanghai, China), and the protein was quantified. Subsequently, the protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to a polyvinylidene fluoride (PVDF) membrane (EMD Millipore Corporation, Billerica, Massachusetts, United States). The cells were immersed in 5% skimmed milk, and cultured with primary antibody ROCK1 (1:1000) at 4°C overnight under the internal reference of β-actin (1:1000). The membrane was washed with TBST (5 min × 3). Subsequently, the cells were cultured with goat anti-mouse/rabbit IgG coupled with horseradish peroxidase (R&D Company, United States) at indoor temperature for 2 h. Finally, the blot of the cells was detected using electrochemiluminescence (ECL) reagent (Thermo Fisher Scientific, United States) and Amersham Prime ECL Plus detection system.
Cell Proliferation Assay
The transfected cells were harvested, and seeded into a 96-well plate at 1×103 cells/well, and 10 μL cell counting kit-8 (CCK8) solution (Dojindo, Japan) was added into each well at each detection time point. After the addition of the solution, the plate was cultured at indoor temperature for 2 h, and then the optimal density of each well at 450 nm was determined using a microplate reader (BioTek Instruments, Inc., Winooski, Vermont, United States).
Cell Invasion Assay and Scratch Adhesion Test
The transwell technology (Corning, United States) was used for cell invasion assay specifically as follows: The cells were transferred to the upper compartment of the Transwell plate coated with Matrigel at 5*104 cells per well. Subsequently, the lower compartment was added with 750 μL FBS, and the plate was incubated for 24 h. After incubation, the invaded cells were immobilized with 4% paraformaldehyde for 30 min, and then rinsed with phosphate buffer saline (PBS). Then, the cells were dyed with 0.1% crystal violet for 15 min, and the number of cells was calculated using a microscope. The cell suspension was transferred to a 6-well plate at 1×106 cells/well, and when the cells reached 90% confluency, the cell layer was scraped with a pipette tip to form a wound. The plate was gently washed with PBS and the cells were incubated for 24 h. Finally, the wound gap was photographed and measured.
Apoptosis Determination
The flow cytometry was adopted to determine cell apoptosis and cell cycle specifically as follows: The cells transfected for 48 h were prepared into cell suspension with 1*106 cells, and the suspension was transferred to a cell culture flask for growth overnight. The collected cells were washed with PBS, and their apoptosis rate was evaluated by an Annexin V-FITC Apoptosis Assay Kit (Invitrogen™, the United States) according to the kit instructions.
Tumor Xenotransplantation
A total of 10 female BALB/c nude mice (4 years old) were collected, and stable MDA-MB-231 cells transfected with sh-NC or sh-LINC01087 were also collected. Then, 2×106 cells from those collected cells were suspended in 100 μL PBS. Afterwards, the suspension was injected into the fourth breast of each nude mouse. The injected mice were feed for 28 days, and their tumor volume was determined using a digital caliper every 7 days as per the formula: Volume = 0.5× width 2× length. After 28 days, the nude mice were euthanized, and then their tumor tissues were sampled for measurement of tumor mass. This study was carried out with permission from the Medical Ethics Committee (Inner Mongolia Key Laboratory of Mongolian Medicine Pharmacology for Cardio-Cerebral Vascular System) of our hospital and in line with the Laboratory animal—Guideline for ethical review of animal welfare GB/T 35892–2018 released by China in 2018.
Statistical Analysis
In this study, the collected data were analyzed statistically and visualized into required figures using GraphPad 7, and the independent prognostic factors of the patients were analyzed using SPSS20.0. The distribution of measurement data was analyzed using the Kolmogorov–Smirnov (K-S) test. Data in normal distribution were expressed as the mean ± standard deviation (Mean±SD), and compared between groups using the independent-samples T-test. Enumeration data were expressed as the percentage (%), and compared using the chi-square test, and expressed by y2. Multi-group comparison was carried out using the one-way ANOVA, and expressed by F. Post hoc pairwise comparison was carried out using the LSD-t-test, and comparison of expression in multiple time points was carried out using the variance of repeated measures, and expressed by F. Post-test was carried out using Bonferroni, and Pearson correlation analysis was carried out to analyze the correlation of genes. The overall survival of the patients was visualized into Kaplan-Meier (K-M) survival curves and studied using the Log rank test. P < 0.05 indicates a significant difference.
Results
High Expression of LINC01087 in Patients with BC Indicates a Poor Prognosis
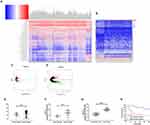
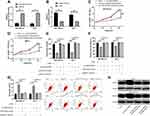
In order to verify the expression of LINC01087 in BC, we first analyzed the expression results of LINC01087 based on TCGA and GEO databases (Figure 1A–D), finding that the LINC01087 was significantly up-regulated in both databases (Figure 1E and F). Then, we determined the expression of LINC01087 in the tissues of patients with BC, and also found high expression of it (Figure 1G). Furthermore, we divided the patients into a high LINC01087 expression group and a low LINC01087 expression group according to the median LINC01087 expression to explore the relationship between LINC01087 and pathological data of the patients. It was found that the high expression group faced significantly higher rates of III+IV stage and lymphatic metastasis and a lower 5-year survival rate than the low expression group (Figure 1H, Table 1).
 |
Table 1 Relationship Between LINC01087 and Pathological Data About the Patients |
Knock-Down of LINC01087 Inhibits the Proliferation, Invasion, and Migration of Breast Cancer Cells (BCCs) and Promotes Apoptosis of Them
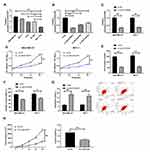
Apart from patients’ BC tissues, we also detected the relative expression of LINC01087 in the patients’ BCCs, and found a significant increase in the expression in the cells (Figure 2A). To verify the influence of LINC01087 on BCC growth, we constructed si-LINC01087#1-#3 interference vectors (Figure 2B), and transfected si-LINC01087#1 interference vectors with the lowest expression of LINC01087 into BCC lines (MDA-MB-231 and MCF-7). It was turned out that transfection of si-LINC01087#1 strongly inhibited the expression of LINC01087 (Figure 2C). We further analyzed the growth of those transfected cells through CCK-8 assay, Transwell assay, scratch adhesion assay, and flow cytometry. The CCK-8 assay revealed that compared with si-NC intervention, si-LINC01087#1 intervention significantly weakened cell proliferation (Figure 2D), and Transwell assay and scratch adhesion assay revealed that si-LINC01087#1 suppressed cell invasion and migration (Figure 2E and F). In addition, flow cytometry revealed that transfection of si-LINC01087#1 dramatically up-regulated cell apoptosis rate (Figure 2G). We also carried out tumor xenotransplantation in nude mice, finding that the volume and mass of tumor in each nude mouse injected with si-LINC01087#1 were significantly smaller than those of tumor in each nude mouse injected with si-NC (Figure 2H). These findings indicated that knock-down of LINC01087 could inhibit BCC growth.
LINC01087 Targetedly Regulates the miR-335-5p Expression in BCCs
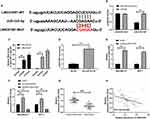
In order to further explore relevant mechanisms of LINC01087, we predicted potential targeted miR of LINC01087 through the online prediction website, http://starbase.sysu.edu.cn/, and found targeted binding locus between LINC01087 and miR-335-5p (Figure 3A). For this reason, we constructed LINC01087-WT and LINC01087-MUT vectors, and carried out a dual-luciferase reporter assay. It was turn out that miR-335-5p-mimics suppressed the fluorescence activity of LINC01087-WT, but did not suppress the activity of LINC01087-MUT, which indicated that LINC01087 could targetedly regulate miR-335-5p (Figure 3B). For the purpose of better verifying the correlation between miR-335-5p and LINC01087, we further carried out RIP assay and RNA pull-down assay. The RIP assay demonstrated that Ago2 antibody precipitated miR-335-5p and LINC01087 more strongly than IgG antibody (Figure 3C), and the RNA pull-down assay revealed that LINC01087 pulled down Bio-miR-335-5p (Figure 3D). We additionally determined the expression of miR-335-5p in cells transfected with si-LINC01087#1 or pcDNA-LINC01087, and found that the miR-335-5p expression in the cells transfected with si-LINC01087#1 significantly increased, while the miR-335-5p expression in those transfected with pcDNA-LINC01087 was reversed (Figure 3E and F). We also found decreased expression of miR-335-5p in BC tissues (Figure 3G), a negative relation between miR-335-5p and LINC01087 (Figure 3H).
MiR-335-5p Suppresses the Invasion and Migration of BCCs by Targetedly Regulating ROCK1
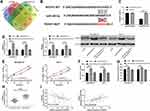
For the purpose of investigating relevant mechanisms of miR-335-5p, we predicted target genes of miR-335-5p through Targetscan, starbase3.0, miRDB, and miRTarBase together, and found a total of 11 target genes (Figure 4A, Table 2). Based on the findings and current references and data, we selected ROCK1 for further analysis, and found through dual-luciferase reporter assay that miR-335-5p could inhibit the activity of ROCK1-WT (Figure 4B and C). In order to verify the ability of miR-335-5p in regulating the BCC growth through ROCK1, we constructed miR-335-5p-mimics+pcDNA-ROCK1, pcDNA-ROCK1, and si-NC, and transfected them into MDA-MB-231 and MCF-7 cells. It was turned out that pcDNA-ROCK1 increased the protein and mRNA expression of ROCK1in cells, while co-transfection of miR-335-5p-mimics and pcDNA-ROCK1 inhibited the increase of ROCK1 expression in the cells (Figure 4D). Cell experiments showed that pcDNA-ROCK1 could intensify cell proliferation (Figure 4E), invasion (Figure 4F), and migration (Figure 4G), while the effects were reversed after co-transfection of it with miR-335-5p-mimics. The experiments also showed that ROCK1 was highly expressed in tissues of the BC patients (Figure 4H), and it was negatively correlated to miR-335-5p, but positively correlated to LINC01087 (Figure 4I).
 |
Table 2 Potential Target Genes Downstream of miR-335-5p Predicted Together |
LINC01087 Regulates the Growth of BCCs via the miR-335-5p/ROCK1 Axis
At the end of this study, we wanted to confirm that LINC01087 could act as a miR-335-5p sponge to affect the biological function of BCCs by regulating ROCK1, so we transfected si-LINC01087#1 + miR-335-5p-inhibit, si-LINC01087#1+pcDNA-ROCK1, si-LINC01087#1, and si-NC into BCCs (MDA-MB-231 and MCF-7) (Figure 5A and B), respectively, and analyzed the biological function of the transfected cells. It was found out that co-transfection of miR-335-5p-inhibit and pcDNA-ROCK1 or si-LINC01087#1 reversed the influence of si-LINC01087#1 on proliferation (Figure 5C and D), invasion (Figure 5E), migration (Figure 5F), and apoptosis (Figure 5G) of BCCs. In addition, we also found that the up-regulation in the relative expression of ROCK1 caused by transfection of miR-335-5p-inhibit and pcDNA-ROCK1 was reversed by co-transfection with si-LINC01087#1 (Figure 5H).
Discussion
In this study, we verified that LINC01087 was overexpressed in BC tissues and could mediate miR-335-5p/ROCK1, and we also found that knock-down of LINC01087 could suppress cell viability, and thus affect the proliferation, invasion, migration, and apoptosis of BCCs, but the addition of miR-335-5p-inhibit and pcDNA-ROCK1 could reverse the effect. It indicated that LINC01087 could be used as a possible therapeutic target for BC and was expected to be a new indicator of it.
LncRNA is a hot research area in recent years. People initially hold that lncRNA was a metabolic “waste” in the transcription process. However, thanks to the continuous improvement of scientific research level this year, growing studies have found that LncRNA is an important part of RNA processing, genome reprogramming, and chromatin modification, and it affects the development and progression of various tumors.20,21 In recent years, many studies have concluded that lncRNA is strongly linked to BC. For example, one study has discovered that LncRNA-ATB enhances the resistance against trastuzumab in BC tissues and intensifies the invasion and metastasis of them,22 and one other study has revealed that LncRNA UCA1 promotes the epithelial-mesenchymal transformation of BCCs by enhancing the Wnt/β-catenin signaling pathway.23 LINC01087 is located on human 2q21.1 chromosome, but there has been no study on it. We analyzed the potential differential lncRNA expression in BC based on TCGA and GEO databases, finding that the expression of LINC01087 in cases with cancer was high in both databases. Therefore, we deduced that LINC01087 may affect the development and progression of BC, so we conducted further research on it.
We first determined the expression of LINC01087 in BC tissues, finding that LINC01087 was highly expressed in BC patients, and patients with high LINC01087 expression faced significantly higher rates of high TNM staging and lymphatic metastasis than those with low LINC01087 expression. We also analyzed the 5-year survival rate of the patients and found that the survival rate of patients with high expression of LINC01087 decreased, which suggested that LINC01087 may affect the development of BC. Moreover, we carried out basic experiments, and found that knock-down of LINC01087 significantly weakened cell viability, in which the proliferation, migration, and invasion of cells were inhibited, while the apoptosis rate was raised. Our tumor xenotransplantation assay also showed that knock-down of it could suppress BCC growth in nude mice, which is consistent with the research results of the study by Tripathi et al.24 These findings indicated that LINC01087 could be a potential target for BC therapy. However, other mechanisms of LINC01087 remain unclear.
ceRNA is an important theory to explain the relationship between lncRNA and miR in recent years.25 Increasing evidences demonstrate that lncRNA acts as a ceRNA to promote changes in the biological function of various tumor cells.26,27 For example, one study by Müller et al28 has found an interaction between lncRNA H19/miR-675 and lncRNA NEAT1/miR-204 in BC, and one other study has revealed that H3K27 acetylation induces the resistance against trastuzumab in BC patients and the epithelial–mesenchymal transition by activating lncRNA TINCR and targeting microRNA-125b.29 In this study, we carried out an online predictive analysis on potential targeted miR of LINC01087 and found that there was binding locus between miR-335-5p and LINC01087. MiR-335-5p, as a tumor suppressor gene, has low expression in many tumors. For the purpose of confirming the relationship between miR-335-5p and LINC01087, we carried out dual-luciferase reporter assay, RIP assay, and RNA pull-down assay on them. It was confirmed that LINC01087 could act as a miR-335-5p sponge to negatively regulate the miR-335-5p expression, and miR-335-5p was lowly expressed in BC patients, and negatively correlated with LINC01087, which indirectly confirmed that LINC01087 could regulate miR-335-5p.
The mode that miR affects tumor growth through targeted regulation of downstream genes has been confirmed by many studies.30 For the purpose of further exploring how miR-335-5p affects the growth of BC, we further predicted its downstream target genes and found a total of 11 potential target genes through the prediction, including ROCK1. ROCK1 is a protein-serine/threonine kinase that is activated when binding to guanosine triphosphate (GTP) binding of Rho and plays a pivotal role in tumor cell invasion and migration.31 For instance, one study has revealed that androgen-regulated miR-135a slows down the migration and invasion of prostate cancer cells by lowering the expression of ROCK1 and ROCK2,32 and one other study has found that miR-145 inhibits the growth and migration of BCCs by targeting oncoprotein ROCK1.33 Based on it, we further verified the correlation of miR-335-5p with ROCK1. Our dual-luciferase reporter assay demonstrated that the luciferase activity of ROCK1-WT could be suppressed by miR-335-5p-mimics, and further experiments demonstrated that miR-335-5p-mimics could inhibit the ROCK1 expression in cells transfected with pcDNA-ROCK1, and it could also reverse the enhancement of pcDNA-ROCK1 on cell proliferation, invasion, and migration, which implied that miR-335-5p could affect the development of BC by targetedly regulating ROCK1. Clinical experiments revealed that ROCK1 was highly expressed in BC, negatively correlated with miR-335-5p, but positively correlated with LINC01087. Therefore, we deduced that LINC01087 can act as a miR-335-5p sponge to affect the expression of ROCK1 and participate in the invasion and migration of BC. In order to verify this point, we co-transfected vectors, and it was turned out that co-transfection of si-LINC01087#1 and miR-335-5p-inhibit or pcDNA-ROCK1 could reverse the effect of si-LINC01087#1 on the proliferation, invasion, migration, and apoptosis of BCCs.
Although this study has confirmed relevant mechanisms of LINC01087 in BC, it still has certain limitations. Firstly, whether LINC01087 can be adopted as a potential diagnostic indicator for BC remains unclear. Secondly, one study by Yang et al34 has found that LINC01087 is lowly expressed in TNBC, but we do not collect patients with TNBC for our study, so we are unable to determine the mechanism of LINC01087 in TNBC. Therefore, we hope to further excavate the function of LINC01087 in BC and the value of it in TNBC to further support our research results.
To sum up, LINC01087 is highly expressed in cases with BC, and patients with high LINC01087 expression suffer a poor survival. Furthermore, LINC01087 can act as a miR-335-5p sponge to regulate the ROCK1 expression and affect the invasion and migration of BCCs.
Conclusion
LINC01087 is highly expressed in BC, and patients with high LINC01087 expression suffer a poor survival. Furthermore, LINC01087 can act as a miR-335-5p sponge to regulate the ROCK1 expression and affect the invasion and migration of BCCs.
Funding
This work was supported by the National Natural Science Foundation of China (No.81873373) and Young Innovative and Entrepreneurial Talents of “Grassland Talents” Project of Inner Mongolia Autonomous Region (No. Q2017042).
Disclosure
The authors report no conflicts of interest in this work.
References
1. DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi:10.3322/caac.21412
2. Jemal A, Robbins AS, Lin CC, et al. Factors that contributed to black-white disparities in survival among nonelderly women with breast cancer between 2004 and 2013. J Clin Oncol. 2018;36(1):14–24. doi:10.1200/JCO.2017.73.7932
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
4. Gray JM, Rasanayagam S, Engel C, Rizzo J. State of the evidence 2017: an update on the connection between breast cancer and the environment. Environ Health. 2017;16(1):94.
5. Howell A, Anderson AS, Clarke RB, et al. Risk determination and prevention of breast cancer. Breast Cancer Res. 2014;16(5):446. doi:10.1186/s13058-014-0446-2
6. Abrahams HJ, Gielissen MF, Schmits IC, Verhagen CA, Rovers MM, Knoop H. Risk factors, prevalence, and course of severe fatigue after breast cancer treatment: a meta-analysis involving 12 327 breast cancer survivors. Ann Oncol. 2016;27(6):965–974. doi:10.1093/annonc/mdw099
7. Lyman GH, Greenlee H, Bohlke K, et al. Integrative therapies during and after breast cancer treatment: ASCO endorsement of the SIO clinical practice guideline. J Clin Oncol. 2018;36(25):2647–2655. doi:10.1200/JCO.2018.79.2721
8. Early Breast Cancer Trialists’ Collaborative G. Adjuvant bisphosphonate treatment in early breast cancer: meta-analyses of individual patient data from randomised trials. Lancet. 2015;386(10001):1353–1361. doi:10.1016/S0140-6736(15)60908-4
9. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi:10.1038/nrc.2017.99
10. Vislovukh A, Vargas TR, Polesskaya A, Groisman I. Role of 3ʹ-untranslated region translational control in cancer development, diagnostics and treatment. World J Biol Chem. 2014;5(1):40–57. doi:10.4331/wjbc.v5.i1.40
11. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi:10.1038/nrc3932
12. Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9. doi:10.1016/j.jbior.2014.09.013
13. Zhou P, Liu P, Zhang J. Long noncoding RNA RUSC1ASN promotes cell proliferation and metastasis through Wnt/beta-catenin signaling in human breast cancer. Mol Med Rep. 2019;19(2):861–868. doi:10.3892/mmr.2018.9763
14. Jing H, Qu X, Liu L, Xia H. A novel long noncoding RNA (lncRNA), LL22NC03-N64E9.1, promotes the proliferation of lung cancer cells and is a potential prognostic molecular biomarker for lung cancer. Med Sci Monit. 2018;24:4317–4323. doi:10.12659/MSM.908359
15. Huo X, Han S, Wu G, et al. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16(1):165. doi:10.1186/s12943-017-0734-4
16. Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi:10.1038/nature20149
17. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286.
18. Edge SB, Compton CC. The American joint committee on cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi:10.1245/s10434-010-0985-4
19. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
20. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
21. Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151–164. doi:10.1016/j.bbcan.2015.07.001
22. Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2016;20(13):2819–2824.
23. Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6(13):11652–11663. doi:10.18632/oncotarget.3457
24. Tripathi R, Aier I, Chakraborty P, Varadwaj PK. Unravelling the role of long non-coding RNA - LINC01087 in breast cancer. Noncoding RNA Res. 2020;5(1):1–10. doi:10.1016/j.ncrna.2019.12.002
25. Haque F, Pi F, Zhao Z, et al. RNA versatility, flexibility, and thermostability for practice in RNA nanotechnology and biomedical applications. Wiley Interdiscip Rev RNA. 2018;9(1):e1452. doi:10.1002/wrna.1452
26. Chen Y, Lin Y, Bai Y, Cheng D, Bi Z. A long noncoding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) network identifies eight lncRNA biomarkers in patients with osteoarthritis of the knee. Med Sci Monit. 2019;25:2058–2065. doi:10.12659/MSM.915555
27. Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148–64167. doi:10.18632/oncotarget.11637
28. Muller V, Oliveira-Ferrer L, Steinbach B, Pantel K, Schwarzenbach H. Interplay of lncRNA H19/miR-675 and lncRNA NEAT1/miR-204 in breast cancer. Mol Oncol. 2019;13(5):1137–1149. doi:10.1002/1878-0261.12472
29. Dong H, Hu J, Zou K, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast cancer. Mol Cancer. 2019;18(1):3. doi:10.1186/s12943-018-0931-9
30. Molotski N, Soen Y. Differential association of microRNAs with polysomes reflects distinct strengths of interactions with their mRNA targets. RNA. 2012;18(9):1612–1623. doi:10.1261/rna.033142.112
31. Sunamura S, Satoh K, Kurosawa R, et al. Different roles of myocardial ROCK1 and ROCK2 in cardiac dysfunction and postcapillary pulmonary hypertension in mice. Proc Natl Acad Sci U S A. 2018;115(30):E7129–E7138. doi:10.1073/pnas.1721298115
32. Kroiss A, Vincent S, Decaussin-Petrucci M, et al. Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene. 2015;34(22):2846–2855. doi:10.1038/onc.2014.222
33. Zheng M, Sun X, Li Y, Zuo W. MicroRNA-145 inhibits growth and migration of breast cancer cells through targeting oncoprotein ROCK1. Tumour Biol. 2016;37(6):8189–8196. doi:10.1007/s13277-015-4722-2
34. Yang F, Liu YH, Dong SY, et al. Co-expression networks revealed potential core lncRNAs in the triple-negative breast cancer. Gene. 2016;591(2):471–477. doi:10.1016/j.gene.2016.07.002
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.