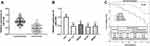Back to Journals » International Journal of General Medicine » Volume 15
LINC01006 and miR-3199 Serve as Novel Markers of Poor Prognosis in Colon Cancer and Regulate Cell Proliferation, Migration and Invasion
Authors Wu Y, Yu B, Li Y, Yu F, Li Z, Chen D, Jiang F, Bo J, Xue H, Lv H, Li H
Received 17 August 2021
Accepted for publication 30 November 2021
Published 16 February 2022 Volume 2022:15 Pages 1677—1687
DOI https://doi.org/10.2147/IJGM.S334701
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Yaoqiang Wu,1 Bo Yu,2 Yaping Li,3 Fuxiang Yu,1 Zhongguo Li,1 Daxin Chen,1 Feng Jiang,1 Jianbo Bo,1 Hongwei Xue,1 Hongyang Lv,1 Haiyang Li1
1Department of General Surgery, Dandong First Hospital, Dandong, Liaoning, People’s Republic of China; 2Department of Gastroenterology, Dandong First Hospital, Dandong, Liaoning, People’s Republic of China; 3Department of Intensive Care Unit, Dandong First Hospital, Dandong, Liaoning, People’s Republic of China
Correspondence: Yaoqiang Wu, Department of General Surgery, Dandong First Hospital, Dandong, Liaoning, People’s Republic of China, Tel/Fax + 86-415-2819133, Email [email protected]
Purpose: Colon cancer is the most commonly diagnosed gastrointestinal cancer. This research intended to evaluate the prognostic values of LINC01006 and miR-3199 for colon cancer and their effects on cell physiology.
Patients and Methods: LINC01006 and miR-3199 expression levels were determined by RT-qPCR. Patients’ 5-year cumulative survival rate was analyzed by Kaplan–Meier curves with the Log rank test. Chi-square test and multivariate Cox regression analysis were used to access the clinical significance. CCK-8 assay, transwell assay, and TUNEL assays were used to monitor the change of cell proliferation, invasion, migration, and apoptosis.
Results: The expression level of LINC01006 was increased while miR-3199 was decreased in colon tissues and cells compared to normal ones. This dysregulated expression was correlated with T stage (P = 0.002) and N stage (P = 0.009). High LINC01006 level (HR = 4.048, 95%: 1.502– 10.911, P = 0.006) or low miR-3199 level (HR = 3.421, 95% CI: 1.254– 9.330, P = 0.016) was outstanding for predicting poor prognosis in patients with colon cancer. Downregulation of LINC01006 reduced cell proliferation, invasion, and migration but induced cell apoptosis (P < 0.05).
Conclusion: LINC01006 knockdown showed anti-proliferative, anti-metastatic, and apoptotic-induced effects on colon cancer cells. This study contributes to research on promising prognostic biomarkers of colon cancer and might give way to further investigation of alternative tumor targets.
Keywords: colon cancer, LINC01006, miR-3199, prognosis
Introduction
With an incidence of more than 1.9 million in 2020, colorectal cancer ranked third among all cancers and is the most diagnosed gastrointestinal cancer.1–3 Colorectal cancers are broadly classified into the colon, rectal and anal cancers.2 Colon cancer has the characteristic of the cancer transition, representing as from infection-related cancer to that linked to rapid societal and economic change.4 Changes in lifestyle and diet, such as high consumption of processed and red meat, high alcohol consumption, and physical inactivity, are steadily increasing the incidence globally.5 Coupled with the introduction of population-based screening and the upgrading treatment, reductions in colon cancer mortality have been observed.3 Nevertheless, survival disparities remain existed, which can partly be attributed to the stage at diagnosis.6 Distant metastases and recurrence render the effect of current treatments and long-term prognosis in advanced cases remains unsatisfactory.7 Therefore, the adoption of effective markers in prognosis would be an important determinant of survival and improve the targeted therapy techniques.
Noncoding RNAs (ncRNAs) are a series of non-protein-coding transcripts, including long noncoding RNAs (lncRNAs, more than 200 nucleotides in length) and microRNAs (miRNAs or miRs, about 20 nucleotides in length).8 As a big surprise of human genome regulatory elements in the postgenomic era, ncRNAs have attracted increasing attention as it is proposed to carry out diverse functions.9–11 ncRNAs are involved in proliferation, differentiation, and apoptosis by various regulations at the cellular level and organismal level.12,13 Given that ncRNAs may function their roles under specific physiological or pathological conditions, ncRNAs show great potential as key molecules in disease, especially cancers.14,15 LINC01006, one kind of long intergenic noncoding RNA (lincRNA), participates in much cellular physiology such as carcinogenesis, proliferation, and metastasis.16–18 Not only LINC01006 could act as functional molecules and regulators for cellular physiology, but also biomarkers in the tumor. A study about biomarkers of gastric cancer suggested LINC01006 might serve as a potent and new prognostic predictor for gastric cancer.19 Another study revealed miR-3199 can be used in building the overall survival-related signature for papillary renal cell carcinoma.20 In colorectal cancer, miR-3199 was screened as a downregulated DEmiRNA.21 As common differentially expressed lncRNA and miRNA in colon cancer,22,23 the functions and functional mechanism of LINC01006 and miR-3199 in colon cancer have not been revealed.
Here, we will detect the expression level of LINC01006 and miR-3199 in colon cancer tissues and cells. Their effects on cellular function would be evaluated. Basic mechanistic will be clarified. Based on this study, we would elucidate the roles of LINC01006 and miR-3199 in clinical use and cellular biology.
Materials and Methods
Patients’ Collection
Medical data of patients who received a pathological diagnosis of colon cancer and have undergone curative surgical resection at Dandong First Hospital were retrospectively reviewed from January 2012 to December 2015. With the inclusion criteria of no neoadjuvant therapy before surgery (inclusive of any radiation-based therapy, chemotherapy, or immunotherapy), a total of 143 patients were selected. The clinical features and 5-year follow-up information of the participants were collected. We reviewed all records associated with colon cancer and recorded tumor stage (AJCC/UICC TNM staging system) and other key parameters (Table 1). Written informed consent was signed by patients who participated. All studies were developed in agreement with the ethics commission (Ethics committee of Dandong First Hospital). This study was also conducted in accordance with the Declaration of Helsinki.
 |
Table 1 Association of LINC01006 and miR-3199 Expression with Key Clinicopathological Characteristics of Patients |
Tissue and Cell Preparation
Human colon cancer tissues and adjacent normal tissues were obtained, preserved in RNAlater (Thermo Fisher Scientific, USA) immediately, and stored at −80°C before use. After disruption using a high-throughput tissue grinder (Scientz-192, China), the RNA in tissues was isolated with TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol.
Normal colon epithelial cell FHC, and four colon carcinoma cell lines HCT116, SW480, HT-29, SW620 cells were used (All from ATCC, USA). SW480 and SW620 cells were cultured in L-15 medium (HyClone, USA) supplemented with 10% FBS (HyClone, China) at an atmosphere of 100% air. FHC, HT-29, and HCT-116 cells were cultured in McCoy’s 5a Medium (Gibco, USA) containing 10% FBS (HyClone, China) and 1% penicillin and streptomycin (Sigma-Aldrich, USA) in a 5% CO2 incubator. Two LINC01006 targeting siRNAs (si-LINC01006-1 and si-LINC01006-2), negative control (si-NC), miR-3199 inhibitor (anti-miR-3199), and miR inhibitor control (anti-NC) were designed and synthesized by Genechem (China). Transient transfections in HCT116 and SW620 cells were performed using the Lipofectamine 2000 (Invitrogen, USA) following the recommended protocol by the manufacturer. Total RNA from cells was obtained using TRIzol reagent (Invitrogen, USA) too.
RT-qPCR Detection for RNA Quantification
The concentration and purity of total RNA were assessed at a NanoDrop 2000 spectrometer (Thermo Fisher Scientific, USA). The relative expression levels of LINC01006 and miR-3199 in the tissues and cells were evaluated by RT-qPCR. Total RNA was used to produce the first-strand cDNA with the use of RevertAid First Strand cDNA Synthesis Kits (Thermo Fisher, USA). PCR amplifications were completed using IQ™ STBR® Green Supermix (Bio-Rad, USA) on a BIO-RAD CFX96 Real-Time PCR Detection System (Bio-Rad, USA). Based on the 2−ΔΔCt method, the relative expression of LINC01006 was normalized to GAPDH, while miR-3199 normalized to U6.
CCK-8 Assay for Cell Proliferation
Cell growth was monitored by using Cell Counting Kit-8 (CCK-8, Dojindo, Japan). As described previously,24 transfected HCT116 and SW620 cells were seeded in 96 well plates, respectively. At the time point of 0, 24, 48, and 72 hours, 10 µL of CCK-8 solution was added to the culture medium and incubated at 37°C for an additional 2 h. The absorbance was detected at the wavelength of 450/630 nm on iMark microplate reader (Bio-Rad, USA).
Transwell Migration and Invasion Assays
The physiological effects of LINC01006 inhibition were examined via Transwell assay. The transwell chambers (BD Biosciences, USA) used for invasion assay were placed into a 24-well plate and then coated with Matrigel, while the transwell chambers for migration assay were not coated. After pre-incubated as confluent monolayers, HCT116 and SW620 cells (1×105 cells/well) were seeded onto serum-starving McCoy’s 5a or L-15 medium in the upper chambers. The lower chambers contained McCoy’s 5a or L-15 medium fortified with 10% FBS for migration assay and 15% FBS for invasion assay. About 24 hours later, cells that migrated or invaded were fixed with 4% PFA and stained with 0.5% crystal violet (Sigma-Aldrich, USA). Cell numbers were obtained under a microscope (Olympus, Japan) in five random fields.
TUNEL Assay for Cell Apoptosis
Apoptosis of HCT116 and SW620 cells with or without LINC01006 downregulation was examined using In Situ Cell Death Detection Kit-Fluorescein (Roche, USA) that based on Tunel method. Briefly, transfected HCT116 and SW620 cells were fixed by 2% PFA and permeated, and then the TUNEL staining was conducted according to the manufacturer’s instructions. The labeled samples were analyzed using Optical microscopy (Olympus, Japan).
Bioinformatic Analysis
To predict the miRNAs targeted to LINC01006, lncRNASNP2, a comprehensive resource of single nucleotide polymorphisms (SNPs) in human lncRNAs, was retrieved (http://bioinfo.life.hust.edu.cn/lncRNASNP#!/).
Dual-Luciferase Reporter Assay
We adapted the Dual-Luciferase reporter assay to verify the binding between LINC01006 and miR-3199. Luciferase reporters containing the wild type LINC01006 3′untranslated region (3′UTR) with a predicted miR-3199 binding site (LINC01006 WT) and mutant LINC01006 3′UTR (LINC01006 MUT), with a mutant region of binding sites, were purchased from Biomart (China). In brief, HCT116 and SW620 cells were cultured in 24-well plates and co-transfected with the indicated luciferase reporter plasmid and anti-miR-3199 or anti-NC using Lipofectamine 2000. The Dual-Glo Luciferase Assay System (Promega, USA) was used to determine the luciferase activities.
Statistical Analysis
Group analysis with two groups was analyzed by t-test. Group analysis with more than two groups was analyzed using a one-way ANOVA, while multiple comparisons used Tukey’s multiple comparisons test. Kaplan–Meier curves were created using the median value of LINC01006 or miR-3199 level as cutoff, and the Log rank test was used to compare overall survival distributions between high-level and low-level groups. The correlation between the expression level of LINC01006 and miR-3199 was analyzed using Pearson correlation analysis. Hazard ratio (HR) of each related parameter was calculated using multivariate Cox regression. P-values of <0.05 were deemed significant.
Results
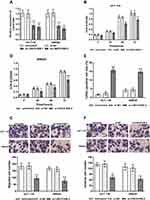
Raised Expression of LINC01006 in Colon Cancer Tissues and Cells Was Associated with Poor Prognosis
To identify LINC01006 showing dysregulated expression in colon cancer, we analyzed RT-qPCR data from the colon cancer tissues and cell lines. As shown in Figure 1A, significant up-regulation was observed in colonic cancerous tissues compared to the para-cancerous normal tissues (P < 0.001). Further, the expression of LINC01006 in intestinal epithelial cells (FHC) and colon cancer cell lines (HCT116, SW480, HT-29, SW620) was examined. Compared with the normal intestinal cells, the cancerous cell lines showed increased expression of LINC01006, which was highest in HCT116 and SW620 cells (P<0.001, Figure 1B). To evaluate the clinical significance of changes in the LINC01006 expression level in colon cancer, the association between the expression level of LINC01006 and the clinicopathological features of the patients were analyzed by Chi-square test. It was identified that LINC01006 expression level was significantly associated with T stage (P=0.002) and N stage (P=0.009) (Table 1), but not significantly with other parameters. Furthermore, the patients were divided into low-level and high-level groups based on the median value of LINC01006 expression. Figure 1C illustrates the lower overall survival rate and shorter overall survival time in patients with high LINC01006 levels when compared with those with low levels through the Kaplan–Meier plot (Log-rank P=0.007). In Table 2, multivariate results from Cox analysis are presented, and the overall multivariate results found that a high LINC01006 level was associated with poorer survival (HR = 4.048, 95%: 1.502–10.911, P=0.006), which is better than traditional biomarkers, including T stage, N stage, Lymphovascular invasion, Extramural venous invasion, Perineural invasion. Combining all these results, the expression level of LINC01006 was increased in colon cancer, and this increase was a significant predictor of overall survival in patients with colon cancer.
 |
Table 2 Significance by Multivariate Analysis of LINC01006 and Other Factors Potentially Associated with Prognosis of Patients with Colon Cancer |
Knockdown of LINC01006 Reduced the Proliferation, Migration, and Invasion, but Induced the Apoptosis of Colon Cancer Cells
After demonstrating an upregulation of LINC01006 in colon cancer, we investigated the effect of LINC01006 knockdown on cell physiology. After validating the successful knockdown of LINC01006 using siRNA (si-LINC01006-1 and si-LINC01006-2, P < 0.001, Figure 2A), we examined the physiological effects after transfection with si-LINC01006-2 via CCK-8 assay, transwell assay, and TUNEL assays. The effect on cell proliferation was analyzed by using CCK-8 assay, and we found that there was a significant reduction in cell proliferation when cells were transfected by LINC01006 siRNA compared to untreated cells (P < 0.05, Figure 2B and C). Next, we performed TUNEL assays and the result suggests the induction of apoptosis in HCT116 and SW620 cells upon LINC01006 downregulation (P < 0.001, Figure 2D). Furthermore, we conducted an analysis of cell migration and invasion using transwell inserts and found LINC01006 knockdown in HCT116 and SW620 cells caused a reduction in cell migratory and invasive ability (P < 0.01, Figure 2E and F). Therefore, LINC01006 plays its role in cell physiology not only by proliferation, migration, and invasion but also by apoptosis.
miR-3199 is Low Expressed in Colon Cancer and Its Expression Correlates with Overall Survival
To identify miR-3199 expression level in colon cancer, we performed RT-qPCR-based RNA expression analysis of human colon cancer tissues and cell lines. We observed that miR-3199 was within the low expression in both colon cancer tissues and cells (P < 0.01, Figure 3A and B). To clarify if the downregulated level of miR-3199 could be clinically relevant, Chi-square test and multivariate Cox regression were employed for correlations and HRs of the involved clinical parameters. The results in Table 1 shows that low miR-3199 level was correlated with T stage (P=0.007), N stage (P=0.028), lymph vascular invasion (P=0.013), and extramural venous invasion (P=0.016). Kaplan–Meier curves (Figure 3C) displayed patients with low miR-3199 levels resulting in poor prognosis (Log-rank P=0.005). By the multivariate analysis (Table 3), a low level of miR-3199 was associated with poor prognostic outcomes (HR = 3.421, 95% CI: 1.254–9.330, P=0.016). Therefore, miR-3199 could be a prognostic factor for colon cancer.
 |
Table 3 Significance by Multivariate Analysis of miR-3199 and Other Factors Potentially Associated with Prognosis of Patients with Colon Cancer |
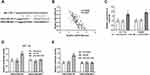
LINC01006 Might Function Its Cellular Role via Binding to miR-3199
Since the effect of LINC01006 on cell physiology has been determined, the regulatory mechanism was then investigated in colon cancer cells. Through bioinformatic analysis in lncRNASNP2, we found that miR-3199 was one of the miRNAs binding to LINC01006 (Figure 4A). Pearson correlation analysis revealed the inverse correlation between the expression level of LINC01006 and that of miR-3199 (P < 0.001, Figure 4B). Then, knockdown of LINC01006 markedly increased miR-3199 level in colon cells from the result of RT-qPCR (P < 0.01, Figure 4C). Besides, the observable increase of relative luciferase activities in cells with wide-type LINC01006 fragment, caused by miR-3199 inhibitor, further validated the interaction of miR-3199 and LINC01006 (P < 0.01, Figure 4D and E).
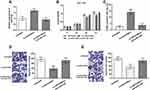
To further confirm that LINC01006 promoted tumor progression through miR-3199 inhibition in colon cancer, functional recovery experiments were carried out in HCT116 cells. As shown in Figure 5A, the transfection was successful (P < 0.001). CCK-8 assay showed the inhibition in cell proliferation caused by LINC01006 knockdown can be restored if miR-3199 was silenced (P < 0.05, Figure 5B). Moreover, the induction of apoptosis in HCT116 cells upon LINC01006 downregulation can be inhibited by miR-3199 silence (P < 0.001, Figure 5C). In addition, miR-3199 silence offset the inhibition of cell migration and invasion upon LINC01006 downregulation (P < 0.01, Figure 5D and E).
Discussion
The significance of ncRNAs has been gradually put under investigation because their aberrant expression is probably a significant factor or a prime mover in the progression of cancer.25 Because lncRNAs and miRNAs often help moderate transcription, they could be optimal biomarkers that are useful in identifying patients at high risk for progression or poor prognosis.26 Numerous discovery studies have shown the promise of ncRNAs as prognostic biomarkers. For instance, lncRNA SNHG12 has been reported as a prognostic indicator for renal cell carcinoma,27 prostate cancer,28 and nasopharyngeal carcinoma.29 LINC01006 is upregulated in cervical cancer and indicated a poor prognosis.16 miR-3199 could serve as a mighty biomarker for recurrence prediction among patients with hepatocellular carcinoma30 and contribute towards survival prediction of lung adenocarcinoma.31 In this study, we detected LINC01006 and miR-3199 in colon cancer tissues and cells at an elevated and a reduced expression level, respectively, via RT-qPCR. Moreover, the high LINC01006 level and the low miR-3199 level were then verified to be associated with a poor prognosis of colon cancer. Based on the results of Chi-square, multivariate Cox regression, and Kaplan–Meier curves, LINC01006 and miR-3199 can serve as independent prognostic factors for patients with colon cancer.
Colon cancer is characterized by uncontrolled cell growth (proliferation), the ability to metastasize to other tissues (migration and invasion), and the absence of the cell death process (apoptosis). Dysregulations in ncRNA expression and subsequent downstream signaling processes have been directly or indirectly implicated in cancer development and progression. Given the dysregulated status of LINC01006 in colon cancer, we performed CCK-8 assay, transwell assay, and cell apoptosis assay with or without LINC01006 knockdown. The results revealed that knockdown of LINC01006 leads to inhibition in cell proliferation, migration, invasion but induction in cell apoptosis. There is a large body of evidence that LINC01006 can affect the cell biology of cancer cells. In pancreatic cancer and prostate cancer, LINC01006 promotes cell growth and metastasis, providing a novel target for cancer therapy.17,18 A recent study about cervical cancer showed LINC01006 downregulation not only inhibited the proliferation and metastasis of cervical cancer cells but also promoted cell apoptosis.17 In this study, LINC01006 exhibits tumor‑promoting functions in colon cancer and provides the basis for identifying LINC01006 as targeted therapy.
LncRNAs exert their functions by multiple mechanisms, in which functioning as decoys and scaffolds of miRNAs, so-called miRNA “sponges”, was the main way.32 LINC01006 has been proved as miR-28-5p sponge in cervical cancer,16 miR-129-2-3p sponge in lung Adenocarcinoma.33 By using bioinformatics analysis and experimental verification, we found LINC01006 was a sponge of miR-3199 in colon cancer cells. After the functional recovery experiments, we infer LINC01006 may exercise its cellular function by binding to miR-3199.
Conclusion
In conclusion, this study has demonstrated the upregulated expression of LINC01006 in colon cancer tissues and cells can promote the progression of cancer, by promoting cell proliferation and metastasis and inhibiting cell apoptosis. LINC01006 knockdown showed anti-proliferative, anti-metastatic, and apoptotic-induced effects in colon cells might be by binding to miR-3199. These findings will contribute to research on promising prognostic biomarkers and might give way to further investigation of alternative tumor targets.
Ethics Statement
Written informed consent was signed by patients who participated. All studies were developed in agreement with the ethics commission (Ethics committee of Dandong First Hospital).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Arnold M, Abnet CC, Neale RE, et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159(1):335–349.e15. doi:10.1053/j.gastro.2020.02.068
3. Araghi M, Soerjomataram I, Jenkins M, et al. Global trends in colorectal cancer mortality: projections to the year 2035. Int J Cancer. 2019;144(12):2992–3000. doi:10.1002/ijc.32055
4. Fidler MM, Bray F, Vaccarella S, Soerjomataram I. Assessing global transitions in human development and colorectal cancer incidence. Int J Cancer. 2017;140(12):2709–2715. doi:10.1002/ijc.30686
5. Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019;11(1):164. doi:10.3390/nu11010164
6. Díaz-Tasende J. Colorectal cancer screening and survival. Revista Espanola de Enfermedades Digestivas. 2018;110(11):681–683. doi:10.17235/reed.2018.5870/2018
7. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi:10.1016/S0140-6736(19)32319-0
8. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36.
9. Palazzo AF, Koonin EV. Functional long non-coding RNAs evolve from junk transcripts. Cell. 2020;183(5):1151–1161.
10. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
11. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465. doi:10.1002/jcp.27486
12. Rinn JL, Chang HY. Long noncoding RNAs: molecular modalities to organismal functions. Annu Rev Biochem. 2020;89:283–308. doi:10.1146/annurev-biochem-062917-012708
13. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi:10.1055/s-0034-1397344
14. Sarfi M, Abbastabar M, Khalili E. Long noncoding RNAs biomarker-based cancer assessment. J Cell Physiol. 2019;234(10):16971–16986. doi:10.1002/jcp.28417
15. Wu Y, Li Q, Zhang R, Dai X, Chen W, Xing D. Circulating microRNAs: biomarkers of disease. Clin Chim Acta. 2021;516:46–54. doi:10.1016/j.cca.2021.01.008
16. Tian L, Han F, Yang J, Ming X, Chen L. Long non‑coding RNA LINC01006 exhibits oncogenic properties in cervical cancer by functioning as a molecular sponge for microRNA‑28‑5p and increasing PAK2 expression. Int J Mol Med. 2021;47(4). doi:10.3892/ijmm.2021.4879
17. Zhang L, Wang Y, Zhang L, et al. LINC01006 promotes cell proliferation and metastasis in pancreatic cancer via miR-2682-5p/HOXB8 axis. Cancer Cell Int. 2019;19:320. doi:10.1186/s12935-019-1036-2
18. Ma E, Wang Q, Li J, Zhang X, Guo Z, Yang X. LINC01006 facilitates cell proliferation, migration and invasion in prostate cancer through targeting miR-34a-5p to up-regulate DAAM1. Cancer Cell Int. 2020;20:515. doi:10.1186/s12935-020-01577-1
19. Zhu X, Chen F, Shao Y, Xu D, Guo J. Long intergenic non-protein coding RNA 1006 used as a potential novel biomarker of gastric cancer. Cancer Biomark. 2017;21(1):73–80. doi:10.3233/CBM-170273
20. Guan Y, Wang B, Zhang T, et al. Integrated analysis revealed the microRNA-based prognostic predicting signature for papillary renal cell carcinoma. DNA Cell Biol. 2021;40(3):532–542. doi:10.1089/dna.2019.5306
21. Dias F, Almeida C, Teixeira AL, Morais M, Medeiros R. LAT1 and ASCT2 related microRNAs as potential new therapeutic agents against colorectal cancer progression. Biomedicines. 2021;9(2):195. doi:10.3390/biomedicines9020195
22. Zhang H, Zhao L, Li S, et al. N6-methylandenosine-related lncRNAs in tumor microenvironment are potential prognostic biomarkers in colon cancer. Front Oncol. 2021;11:697949. doi:10.3389/fonc.2021.697949
23. Ma R, Zhao Y, He M, et al. Identifying a ten-microRNA signature as a superior prognosis biomarker in colon adenocarcinoma. Cancer Cell Int. 2019;19:360. doi:10.1186/s12935-019-1074-9
24. Liao X, Chen J, Liu Y, et al. Knockdown of long noncoding RNA FGFR3- AS1 induces cell proliferation inhibition, apoptosis and motility reduction in bladder cancer. Cancer Biomark. 2018;21(2):277–285. doi:10.3233/CBM-170354
25. Wang J, Song YX, Ma B, et al. Regulatory roles of non-coding RNAs in colorectal cancer. Int J Mol Sci. 2015;16(8):19886–19919. doi:10.3390/ijms160819886
26. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179(5):1033–1055. doi:10.1016/j.cell.2019.10.017
27. Liu Y, Cheng G, Huang Z, et al. Long noncoding RNA SNHG12 promotes tumour progression and sunitinib resistance by upregulating CDCA3 in renal cell carcinoma. Cell Death Dis. 2020;11(7):515. doi:10.1038/s41419-020-2713-8
28. Cheng G, Song Z, Liu Y, et al. Long noncoding RNA SNHG12 indicates the prognosis of prostate cancer and accelerates tumorigenesis via sponging miR-133b. J Cell Physiol. 2020;235(2):1235–1246. doi:10.1002/jcp.29039
29. Liu ZB, Tang C, Jin X, Liu SH, Pi W. Increased expression of lncRNA SNHG12 predicts a poor prognosis of nasopharyngeal carcinoma and regulates cell proliferation and metastasis by modulating Notch signal pathway. Cancer Biomark. 2018;23(4):603–613. doi:10.3233/CBM-181873
30. Bai F, Zhou H, Ma M, Guan C, Lyu J, Meng QH. A novel RNA sequencing-based miRNA signature predicts with recurrence and outcome of hepatocellular carcinoma. Mol Oncol. 2018;12(7):1125–1137. doi:10.1002/1878-0261.12315
31. Yerukala Sathipati S, Ho SY. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci Rep. 2017;7(1):7507. doi:10.1038/s41598-017-07739-y
32. Kazimierczyk M, Kasprowicz MK, Kasprzyk ME, Wrzesinski J. Human long noncoding RNA interactome: detection, characterization and function. Int J Mol Sci. 2020;21(3):1027. doi:10.3390/ijms21031027
33. Zhang Y, Liu H, Zhang Q, Zhang Z. Long noncoding RNA LINC01006 facilitates cell proliferation, migration, and epithelial-mesenchymal transition in lung adenocarcinoma via targeting the microRNA 129-2-3p/CTNNB1 axis and activating Wnt/β-catenin signaling pathway. Mol Cell Biol. 2021;41(6):e0038020. doi:10.1128/MCB.00380-20
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.