Back to Journals » Cancer Management and Research » Volume 12
LINC00963 Confers Oncogenic Properties in Glioma by Regulating the miR-506/BCAT1 Axis
Authors Ye F, Xu R, Ge Y, Zheng Y, Liu X, Deng P, Xu X
Received 16 January 2020
Accepted for publication 16 March 2020
Published 27 March 2020 Volume 2020:12 Pages 2339—2351
DOI https://doi.org/10.2147/CMAR.S246332
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Feng Ye, Ronghua Xu, Yuanhong Ge, Yi Zheng, Xiaowei Liu, Pingfu Deng, Xuejun Xu
Department of Neurosurgery, The Second People’s Hospital of Chengdu, Chengdu, Sichuan 610021, People’s Republic of China
Correspondence: Xuejun Xu
Department of Neurosurgery, The Second People’s Hospital of Chengdu, 10 Qingyun South Street, Chengdu, Sichuan 610021, People’s Republic of China
Email [email protected]
Background: Glioma is a prevalent disease of the central nervous system with a high incidence and mortality rate. Many long noncoding RNAs (lncRNAs) have been determined to be critical regulators of glioma oncogenesis. However, the function and mechanism of LINC00963 in glioma have not been fully elucidated.
Methods: The expression level of RNA was determined by qRT-PCR, and the protein level was determined by Western blot analysis. A luciferase activity assay was conducted to verify the interaction between miRNA and lncRNA or the target gene. The proliferation, cell cycle distribution, invasion, and migration were evaluated by MTT, EdU, flow cytometry, wound-healing and Transwell invasion assays, respectively. In vivo tumor growth was evaluated in a xenograft nude mouse model.
Results: We found that LINC00963 was upregulated in glioma cells and tissues and associated with the poor prognosis of patients with glioma. Ectopic expression of LINC00963 promoted cell proliferation, cell cycle progression, migration, and invasion in vitro and tumorigenesis in vivo. Mechanistically, the results of luciferase activity and RNA pulldown assays validated that LINC00963 could act as a molecular sponge of miR-506. Reciprocal repression was found between LINC00963 and miR-506. In addition, BCAT1 was identified as a target of miR-506, and both the mRNA and protein levels of BCAT1 were reduced by miR-506. In tumor tissues, the expression of BCAT1 was negatively and positively correlated with miR-506 and LINC00963 expression, respectively. The reintroduction of BCAT1 in glioma cells abolished the tumor suppressive function of miR-506 by promoting cell viability and motility. The upregulated LINC00963 and BCAT1 were associated with the aggressive phenotypes of tumors.
Conclusion: Our data revealed that LINC00963 confers oncogenic function in the progression of glioma and that the LINC00963/miR-506/BCAT1 axis may be a novel mechanism and therapeutic strategy for this disease.
Keywords: LINC00963, miR-506, BCAT1, glioma, ceRNA, invasion
Introduction
Long noncoding RNAs (lncRNAs) are defined as a large group of transcripts longer than 200 nucleotides with limited or no potential for protein coding.1 Recently, an increasing number of studies have shown that lncRNAs are frequently dysregulated in various human malignancies and implicated in cancer progression.2 Several lncRNAs, such as MALAT1, XIST, and NEAT1, have been determined to be involved in the development of glioma,3–5 suggesting the utility of lncRNAs as diagnostic markers and therapeutic targets for this disease. In addition to lncRNAs, microRNAs (miRNAs) are noncoding RNAs (ncRNAs) that play pivotal roles in tumor-related cellular processes.6 Increasing evidence suggests that lncRNAs regulate the expression of mRNAs and act as competing endogenous RNAs (ceRNAs) by binding miRNAs. For example, HOXA11-AS could promote renal cancer cell growth and invasion by regulating the miR-146b-5p/MMP16 axis.7 GAS5 acts as a ceRNA for miR-21 to stimulate the expression of BAX and promote apoptosis in laryngeal squamous cell carcinoma.8 TUG1 promoted cervical cancer malignant progression and activated SIRT1 and the downstream Wnt/β-catenin signaling pathway by sponging miR-138-5p.9
Glioma is one of the most aggressive malignant tumors in the human central nervous system, with a high mortality rate being observed worldwide.10 The underlying molecular mechanisms of glioma tumorigenesis has not been elucidated. Therefore, verifying novel and pivotal molecules involved in the development of glioma may provide insights for understanding and treating glioma. Many studies have found that abnormal expression of lncRNAs and miRNAs plays a critical role in glioma progression.11,12 The ceRNA network was also observed in glioma. For example, CRNDE contributes to cell proliferation, migration, and invasion by sponging miR-384 to PIWIL4 expression.13 SNHG16 could increase the expression of the miR-4518-targeted gene PRMT5 by functioning as a sponge of miR-4518, thereby promoting cell viability and inducing apoptosis.14
Among these dysregulated lncRNAs, LINC00963 was reported to be related to the progression of various types of cancers. Knockdown of linc00963 attenuated proliferation, motility, invasion ability, EGFR expression and AKT phosphorylation levels and promoted cell apoptosis.15 LINC00963 predicts poor prognosis in patients with melanoma, and LINC00963 silencing inhibits cell proliferation, migration and invasion by interacting with miR-608.16 LINC00963 was observed to be highly expressed in osteosarcoma tissues and cells, and LINC00963 overexpression was found to promote cell viability and motility in a miR-204-3p-dependent manner.17 However, the function and mechanism of LINC00963 in glioma have not been thoroughly elucidated.
In this study, we aimed to study the involvement of LINC00963 in glioma and investigate its function and mechanism during the progression of glioma. The results demonstrated that LINC00963 effectively acted as a sponge for miR-506 to activate the expression of BCAT1, ultimately leading to an increase in cell growth and metastasis.
Materials and Methods
Patients and Tissue Specimens
We collected 50 pairs of glioma tissues and matched adjacent nontumor tissues from patients who underwent surgical resection at the Second People’s Hospital of Chengdu from Jan 2015 to Dec 2016. All samples were histopathologically diagnosed by pathologists and classified according to the 2007 WHO classification. All patients did not receive chemotherapy or radiotherapy prior to collection. Samples were immediately frozen in liquid nitrogen and stored at −80 °C until use. All patients provided written informed consent, and that this was conducted in accordance with the Declaration of Helsinki. This study was approved by the ethics committee of Second People’s Hospital of Chengdu.
Cell and Transfection
Human glioma cell lines (U87, U251, SHG44, A172) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China), as well as normal human astrocytes (NHAs). Cells were cultured with RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 100 U/mL penicillin/streptomycin (Invitrogen) in a humidified incubator with 5% CO2 at 37 °C.
To construct LINC00963-overexpressing cells, the full-length human LINC00963 sequence was amplified by PCR, and the product was subcloned into a pcDNA3.1 vector (Invitrogen) and named pcLINC00963. Specific siRNAs were designed to knockdown LINC00963 (si-RNA1, 5ʹ-GGTTCCTCATCTGCCAGTT-3ʹ; si-RNA2, 5ʹ-GGCGCAGTAACAATATAAT-3ʹ), and si-NC was used as negative control. A negative control (pcNC) was also constructed. MiR-506 mimic, miR-506 inhibitor and their corresponding negative controls (miR-NC and inh-NC) were purchased from GenePharma (Shanghai, China). BCAT1-expressing vectors (named pcBCAT1) were generated by GenePharma and used to restore the expression of BCAT1 in cells, and pcDNA was used as a control. The transfection procedure was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen). After being reverse-transcribed to cDNA using a PrimeScript RT reagent Kit (Takara, China), qRT-PCR assay was performed by Light Cycler 480 Mix (Roche, Germany) in a total volume of 20 µL on a LightCycler 480 PCR system (Roche). The mRNA expression was normalized to GAPDH, and miRNA and lncRNA expression was relative to U6. The relative expression was calculated using the 2−ΔΔCt method. The following primers were used: LINC00963, forward: 5ʹ-CCACTCTGCTACTGACCACA-3ʹ and reverse: 5ʹ-TCAAGACACCACAGCAGCTA-3ʹ; miR-506, forward: 5ʹ-TCCGAAACGGGAGAGTTGG-3ʹ and reverse: 5ʹ-GTGCAGGGTCCGAGGT-3ʹ; BCAT1, forward: 5ʹ-AGCCCTGCTCTTTGTACTCTT-3ʹ and reverse: 5ʹ-CCAGGCTCTTACATACTTGGGA-3ʹ. All samples were analyzed in triplicate.
Cell Proliferation and Flow Cytometry Assays
The viability of cells at 24, 48, 72 and 96 h was detected by MTT assay. Briefly, cells were cultured in 96-well plates, and 20 μL of MTT (5 mg/mL, Sigma, St. Louis, MO, USA) was added to each well and incubated for 4 h. The absorbance at 490 nm was measured by a microplate reader.
For cell cycle analysis, cells were collected, washed, and fixed in 70% cold ethanol overnight. A total of 1×106 cells were resuspended in propidium iodide (PI, 50 μg/mL, Sigma)/RNase buffer (BD Biosciences, San Jose, CA, USA), and cells were incubated for 30 min at 37°C in the dark. The cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences).
EdU Staining
EdU staining was conducted with a BeyoClick™ Cell Proliferation Kit (Beyotime, Shanghai, China). Upon addition of fresh medium, EdU was added and cells were incubated for 2 h. After incubation, cells were washed by PBS and immobilized for 30 min in PFA (Sigma) before being stained by DAPI (Sigma). Following an additional wash with PBS, cells were observed via an inverted microscope (Olympus, Tokyo, Japan).
Wound-Healing and Transwell Invasion Assays
For the wound-healing assay, transfected cells were seeded into 24-well plates at a density of 1×105 cells per well and incubated overnight. A 10-μL pipette tip was used to create the wound in the center of the plate, and the plate was washed with PBS 3 times to remove the floating cells. Cell movement was recorded at 0 and 24 h by Image-Pro Plus 6.0 software.
A 24-well Transwell insert (Corning, Corning, NY, USA) precoated with Matrigel and with 8-µm pores was used to determine the invasive ability of cells. Cells (2×105) in 100 µL serum-free medium were seeded in the upper chamber, and 400 µL medium with 10% FBS was added into the lower chamber. After 24 h incubation, the bottom cells were fixed in methanol, stained with 0.1% crystal violet, photographed and counted under an inverted microscope (Olympus, Japan).
Dual Luciferase Activity Assay
To reveal the relationship between LINC00963 and miR-506 as well as miR-506 and BCAT1, the seed sequence of BCAT1 3ʹ-UTR or LINC00963 was amplified by PCR and then individually inserted into the pmiRGLO reporter vector (Promega, Madison, WI, USA) to generate wild-type LINC00963 reporter (LINC00963-wt) and BCAT1 reporter (BCAT1-wt). The mutant LINC00963 reporter (LINC00963-mut) and BCAT1 reporter (BCAT1-mut) were generated using a fast site-directed mutagenesis kit (Tiangen, Beijing, China). The combined plasmids LINC00963-wt (or LINC00963-mut) were cotransfected into glioma cells accompanied by miR-506 mimic (or miR-NC) using Lipofectamine 2000 (Invitrogen), which is similar to the cotransfection of BCAT1 plasmids and miR-506 mimic. Luciferase activity was measured 48 h posttransfection using the Dual-Luciferase Reporter Assay System (Promega).
Pulldown Assay with Biotinylated miR-506
Cells were transfected with biotin-labeled wild-type (wt) miR-506 (Bio-miR-506-wt), mutated (mt) miR-506 (Bio-miR-506-mt) or antagonistic miR-506 probe (Bio-NC). Two days after transfection, cells were harvested and incubated with a specific lysate buffer (Ambion, Austin, TX, USA) for 10 min, and cell lysates were mixed with M-280 streptavidin magnetic beads (Sigma) for 3 h at 4 °C. The pulldown products were subjected to qRT-PCR assay.
Western Blot Analysis
Protein was extracted using RIPA Reagent (Beyotime, Beijing, China) and quantified by a BCA protein assay kit (Beyotime). A total of 20 µg protein were separated by SDS-PAGE and transferred onto a PVDF membrane (Millipore, Boston, MA, USA). The membrane was blocked with fat-free milk for 2 h at room temperature and then incubated with primary antibodies at 4 °C overnight. The first antibody included BCAT1 (1:1000 dilution) (Cell Signaling Tech., Danvers, MA, USA). After being washed with TBST, the membrane was then incubated with the secondary antibody for 2 h. Protein bands were visualized by an ECL chemiluminescence kit (Millipore).
Tumor Xenograft Model
Ten female athymic BALB/c nude mice (4–6 weeks old) were obtained from Shanghai SLAC Laboratory Animal Center (Shanghai, China). This study was approved by the Animal Ethics Committee of the Second People’s Hospital of Chengdu. Meanwhile. We confirmed that all animal procedures were performed in accordance with the National Guidelines for Experimental Animal Welfare (the Ministry of Science and Technology, China). U87 cells (5×106) stably transfected with LINC00963 expressing vector (pcLINC00963) or empty vector (pcNC) were subcutaneously injected into the flanks of nude mice (n=5 per group). The tumor volumes were measured every 5 days and calculated using the following formula: volume = length×width2×0.5. After 32 days of injection, all mice were sacrificed, and mouse tumors were obtained for further experiments, including immunohistochemistry (IHC) for Ki-67.
Statistical Analysis
Statistical analyses were performed by SPSS 16.0 (IBM, Chicago, IL, USA.) and GraphPad Prism 5.0 (GraphPad, CA, USA). Data were presented as means±standard deviation (SD). The significance of differences among groups was estimated by Student’s t-test or one-way ANOVA. The overall survival (OS) of the cases was calculated with the Kaplan-Meier method and Log rank test. P<0.05 was considered to be significant.
Results
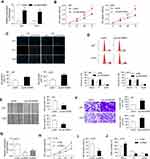
LINC00963 Was Increased in Glioma Cells and Tissues
First, LINC00963 was significantly upregulated in glioblastoma multiforme tissues according to the data from GEPIA (http://gepia.cancer-pku.cn/index.html) (Figure 1A). We also evaluated the expression of LINC00963 in glioma cases in our study. The results of the qRT-PCR assay showed that LINC00963 was overexpressed in glioma cells compared to normal control cells (P<0.01, Figure 1B). LINC00963 expression was significantly higher in tumor tissues (0.349±0.120) than in adjacent noncancerous tissues (0.195±0.089, P<0.001, Figure 1C). The mean level of LINC00963 expression in tumors was used as the cut-off value to divide cases into high or low groups. The high expression of LINC00963 indicated poor prognosis for patients (P=0.009, Log rank test, Figure 1D).
LINC00963 Contributed to Cell Growth and Invasion
To determine the biological function of LINC00963 in glioma cells, we performed gain-of function analysis. The transfection efficiency was determined by qRT-PCR, and the results showed that LINC00963 expression was significantly increased in glioma cells after transfection with pcLINC00963 vectors (Figure 2A). The MTT assay showed that cell proliferation was markedly increased in LINC00963-transfected cells (Figure 2B). EdU positive cells were increased due to LINC00963 overexpression (Figure 2C).
Overexpression of LINC00963 led to an increase in cells at S phase and a decrease in cells at G0/G1 phase (Figure 2D). In addition, the results of wound-healing and Transwell invasion assays showed that LINC00963 facilitated cell migratory and invasive capability (Figure 2E and F). A172 cells, which express relatively high expression of LINC00963, was used to knockdown of LIN00963. As shown in Figure 2G, the expression of LINC00963 was effectively inhibited by si-RNA1, which was used in further experiments. Knockdown of LINC00963 significantly decreased cell proliferation rate, EdU positive cells, and the number of cells at S phase (Figure 2H–J). These results demonstrated that LINC00963 might act as an oncogene in glioma development.
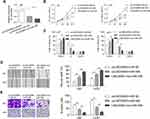
Interaction Between LINC00963 and miR-506
To study the possible ceRNA network involving LINC00963, we used bioinformatic tools (Starbase 3.0) and found that miR-506 has potential binding sites on LINC00963 (Figure 3A). The miR-506 mimic reduced the luciferase activity of the reporter vector containing wild-type LINC00963 but not the mutant type (Figure 3B). The miR-506 mimic control had no influence on the luciferase activity of either wild-type or mutant LINC00963 (Figure 3B). Moreover, an RNA pulldown assay revealed that LINC00063 was coprecipitated with Bio-miR-506-wt rather than Bio-miR-506-mt (Figure 3C). The expression of miR-506 was decreased by LINC00963 overexpression in U87 and U251 cells, while increased by LINC00963 silencing in A172 cells (Figure 3D and E). Conversely, the expression of LINC00963 was inhibited in miR-506 mimic-transfected cells compared to control cells (Figure 3F). We also measured the expression of miR-506 in clinical samples and found that miR-506 was significantly downregulated in glioma tissues compared to normal control tissues (0.205±0.049 vs. 0.543±0.236, P<0.001, Figure 3G) and negatively correlated with LINC00963 expression in tumors (Pearson r=−0.314, P=0.027, Figure 3H). These data suggested that miR-506 was a target of LINC00963 in glioma.
miR-506 Reversed the Oncogenic Function of LINC00963
Whether miR-506 involve in LINC00963 regulated tumorigenesis were investigated in in glioma cells. Cells were divided into three groups: pcLINC00963 plus miR-NC, pcLINC00963 plus miR-506, and LINC00963-mut plus miR-506. The transfection efficiency were determined by qRT-PCR assay (Figure 4A). We found that miR-506 significantly abolished the LINC00963 induced cell proliferation and cell cycle progression (Figure 4B and C). Cell growth was significantly inhibited in LINC00963-mut plus miR-506 group when compared with pcLINC00963 plus miR-506 group (Figure 4B and C). Likewise, miR-506 reversed cell migration and invasion induced by LINC00963 (Figure 4D and E). However, mutation of LINC00963 failed to neutralization the suppressive effect of miR-506 on cell motility (Figure 4D and E). These data validated that miR-506 was a functional target of LINC00963 in glioma.
BCAT1 Is a Direct Target of miR-506
Three different databases (targetScan, miRDB, and picTar) were used to search for potential targets of miR-506. Among these genes, BCAT1 was reported to be related to the progression of human cancer, and the expression of BCAT1 was increased in glioblastoma multiforme based on GEPIA (Figure 5A). MiR-506 has potential binding sites on the 3ʹ-UTR of BCAT1 mRNA (Figure 5B). A luciferase reporter assay indicated that miR-506 significantly reduced the luciferase activity of wild-type BCAT1 reporters but not the mutant reporters (Figure 5C). Both the mRNA and protein levels of BCAT1 were reduced by the miR-506 mimic but increased by the miR-506 inhibitor (Figure 5D and E). LINC00963 elevated the mRNA and protein expression of BCAT1 in glioma cells (Figure 5F and G). Besides, silencing of LINC00963 reduced the mRNA and protein expression of BCAT1 in A172 cells (Figure 5H and I). These data suggested that the LINC00963-regulated ceRNA network may exist in glioma progression.
BCAT1 Was Associated with Glioma Progression
The mRNA levels of BCAT1 were dramatically increased in tumor tissues compared with normal control tissues (1.730±0.376 vs. 0.921±0.253, P=0.0003, Figure 6A). The expression of BCAT1 was inversely and positively correlated with miR-506 and LINC00963, respectively (Pearson r=−0.442 and 0.375, respectively, Figure 6B and C). These data indicated that BCAT1 was a direct target of miR-506 and could be regulated by LINC00963.
In addition, we also evaluated the influence of dysregulated LINC00963/miR-506/BCAT1 on clinicopathological features and patient overall survival. The expression of genes below and above the mean value in tumors was defined as the low- and high-expression groups, respectively. We found that high expression of LINC00963 and BCAT1 was significantly associated with the advanced stage of tumors (P=0.002 and 0.029, respectively, Table 1). Interestingly, the high expression of LINC00963 indicated more elderly patients (P=0.036). By using univariate Cox regression analysis, we found that both higher TNM stage and high levels of LINC00963 were associated with a significantly increased risk of death (HR=3.649 and 3.085, respectively, Table 2). However, only TNM stage was confirmed as a nearly independent prognostic factor by multivariate analysis (Table 2).
 |
Table 1 Correlations Between Clinicopathological Features and Target Gene Expression in 50 Patients with Glioma |
 |
Table 2 Cox Proportional Hazards Regression Analysis |
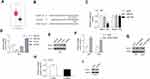
Biological Function of miR-506 Was Partially Reversed by BCAT1
We also evaluated the effect of miR-506 and BCAT1 on cellular processes. The expression of BCAT1 mRNA and protein was determined after cotransfection of miR-506 (or miR-NC) and pcBCAT1 (or pcDNA) in U87 cells (Figure 7A and B). Functionally, restoration of BCAT1 abolished the miR-506-induced inhibition of cell proliferation and cell cycle transition (Figure 7C and D). Likewise, ectopic expression of miR-506 attenuated cell migration and invasion, which could be recovered by BCAT1 (Figure 7E and F). These data suggested that miR-506 inhibited the aggressive tumor phenotype, at least in part, by interacting with BCAT1.
LINC00963 Promoted Tumor Growth in vivo
The effect of LINC00963 on tumor growth was also evaluated by in vivo analysis. Stably overexpressing LINC00963 induced a significantly larger tumor size (Figure 8A and B). The tumor wights were higher in LINC00963-treated mice than in control mice (Figure 8C). Moreover, the proliferative marker Ki-67 was noticeably enhanced in the LINC00963 treatment group (Figure 8D). These results suggest that LINC00963 confers oncogenic function in the development of glioma.
Discussion
Increasing studies have shown that lncRNAs play an essential role in tumor progression. In our study, the expression of LINC00963 was upregulated in glioma cells and tissues, suggesting that LINC00963 may have important roles in the development of glioma. Then, by gain-of-function analysis, we found that LINC00963 acted as an oncogenic lncRNA by promoting cell proliferation, cell cycle progression, migration, and invasion in vitro, which was similar to the findings of previous studies in other cancers.16,17 LINC00963 contributes to tumorigenesis and radioresistance by activating ACK1 expression in breast cancer.18 Rescue of LINC00963 abolished the inhibitory effect of miR-1193 on cell proliferation and migration in cutaneous squamous cell carcinoma.19 In this study, we found that LINC00963 facilitated cell cycle progression from G0/G1 phase to S phase. Similarly, overexpression of LINC00963 enhanced the proliferation ability of hepatocellular carcinoma cells by increasing the number of cells at G0/G1 phase.20 Clinically, the high expression of LINC00963 was associated with tumor stage and prognosis, which was similar to the findings of previous studies.16,18,19 All these results revealed that LINC00963 might be a critical promoter in oncogenesis, as well as in glioma.
It has been widely accepted that lncRNAs can combine with miRNAs to improve tumor progression. Among the predicted miRNAs, we focused on miR-506 based on previous studies showing that miR-506 is involved in the biological processes of glioma.21,22 Then, the results of luciferase activity and RNA pulldown assays led us to conclude that miR-506 is a target gene of LINC00963. LINC00963 and miR-506 were negatively regulated by each other in glioma cells, and they were inversely correlated in tumor tissues. Our data revealed a novel miRNA target of LINC00963 in the pathogenesis of glioma. Certainly, several other miRNAs were identified as targets of LINC00963 in different cancers. For example, miR-608, miR-204-3p, miR-324-3p, and miR-1193 were experimentally validated as targets of LINC00963 in various types of human tumors.16–19 These data suggested that LINC00963 might exert an oncogenic role in different cancers by sponging different miRNAs.
Meanwhile, our data indicated the tumor suppressor role of miR-506 in glioma pathogenesis owing to its suppressive effect on cell growth, migration, and invasion, which was similar to previous studies.21,22 Furthermore, miR-506 inhibits cell proliferation, adhesion, and invasion in breast cancer cells by inactivating IQGAP1 and its downstream ERK/MAPK signaling pathways.23 Ectopic expression of miR-506 suppressed epithelial-to-mesenchymal transition and angiogenesis in gastric cancer.24 Depletion of miR-506 promoted apoptosis in retinoblastoma cell lines.25 MiR-506 was also found to interact with other lncRNAs, such as NEAT1 and MALAT1, to regulate cancer biological processes.26–28 Thus, we suggest that miR-506 might be sponged by multiple oncogenic lncRNAs to affect tumorigenesis. It would be possible to mediate a target gene of miR-506 if LINC00963 acts as a ceRNA in glioma. In our study, BCAT1 was predicted and experimentally validated as a target of miR-506. MiR-506 reduced the expression of BCAT1 at both the mRNA and protein levels. Moreover, the expression of BCAT1 was induced by LINC00963 in glioma cells, supporting the ceRNA regulatory mechanism between them. BCAT1 was involved in miR-506-regulated cellular behavior by promoting cell growth, migration, and invasion, suggesting the oncogenic role of BCAT1 in glioma. We found significantly overexpressed BCAT1 mRNA levels in glioma tissues, which was in keeping with the data from the Oncomine database (data not shown). Suppression of BCAT1 in glioma cells resulted in decreased proliferation and invasiveness in vitro and tumor growth in vivo.29 BCAT1 indicated poor prognosis and led to cell proliferation and chemoresistance in hepatocellular carcinoma.30 BCAT1 could be activated by c-Myc, and overexpression of BCAT1 induces cell proliferation, migration and invasion in nasopharyngeal carcinoma.31 In total, BCAT1 may be a pivotal oncogene in human malignancies, including glioma.
Conclusion
Our study demonstrated the important role of the LINC00963/miR-506/BCAT1 regulatory axis in the cell growth and metastasis of glioma, providing potential targets and strategies to understand the mechanism of glioma carcinogenesis.
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46.
2. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
3. Cao S, Wang Y, Li J, Lv M, Niu H, Tian Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am J Cancer Res. 2016;6(11):2561–2574.
4. Yao Y, Ma J, Xue Y, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359(1):75–86. doi:10.1016/j.canlet.2014.12.051
5. Zhou K, Zhang C, Yao H, et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17(1):105. doi:10.1186/s12943-018-0849-2
6. Beyer S, Fleming J, Meng W, Singh R, Haque SJ, Chakravarti A. The role of miRNAs in angiogenesis, invasion and metabolism and their therapeutic implications in gliomas. Cancers (Basel). 2017;9(7):E85.
7. Yang FQ, Zhang JQ, Jin JJ, et al. HOXA11-AS promotes the growth and invasion of renal cancer by sponging miR-146b-5p to upregulate MMP16 expression. J Cell Physiol. 2018;233(12):9611–9619. doi:10.1002/jcp.26864
8. Lyu K, Xu Y, Yue H, et al. Long noncoding RNA GAS5 acts as a tumor suppressor in laryngeal squamous cell carcinoma via miR-21. Cancer Manag Res. 2019;11:8487–8498. doi:10.2147/CMAR.S213690
9. Zhu J, Shi H, Liu H, Wang X, Li F. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017;8(39):65253–65264. doi:10.18632/oncotarget.18224
10. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi:10.1056/NEJMra0708126
11. Zhou Q, Liu J, Quan J, Liu W, Tan H, Li W. MicroRNAs as potential biomarkers for the diagnosis of glioma: a systematic review and meta-analysis. Cancer Sci. 2018;109(9):2651–2659. doi:10.1111/cas.13714
12. Rynkeviciene R, Simiene J, Strainiene E, et al. Non-Coding RNAs in Glioma. Cancers (Basel). 2018;11(1):17. doi:10.3390/cancers11010017
13. Zheng J, Liu X, Wang P, et al. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24(7):1199–1215. doi:10.1038/mt.2016.71
14. Lu YF, Cai XL, Li ZZ, et al. LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in glioma. Cell Physiol Biochem. 2018;45(5):1975–1985. doi:10.1159/000487974
15. Wang L, Han S, Jin G, et al. Linc00963: a novel, long non-coding RNA involved in the transition of prostate cancer from androgen-dependence to androgen-independence. Int J Oncol. 2014;44(6):2041–2049. doi:10.3892/ijo.2014.2363
16. Jiao H, Jiang S, Wang H, Li Y, Zhang W. Upregulation of LINC00963 facilitates melanoma progression through miR-608/NACC1 pathway and predicts poor prognosis. Biochem Biophys Res Commun. 2018;504(1):34–39. doi:10.1016/j.bbrc.2018.08.115
17. Zhou Y, Yin L, Li H, Liu LH, Xiao T. The LncRNA LINC00963 facilitates osteosarcoma proliferation and invasion by suppressing miR-204-3p/FN1 axis. Cancer Biol Ther. 2019;20(8):1141–1148. doi:10.1080/15384047.2019.1598766
18. Zhang N, Zeng X, Sun C, et al. LncRNA LINC00963 promotes tumorigenesis and radioresistance in breast cancer by sponging miR-324-3p and inducing ACK1 expression. Mol Ther Nucleic Acids. 2019;18:871–881. doi:10.1016/j.omtn.2019.09.033
19. Wang J, Li C, Xu L, Yang C, Zhang X. MiR-1193 was sponged by LINC00963 and inhibited cutaneous squamous cell carcinoma progression by targeting SOX4. Pathol Res Pract. 2019;215(10):152600. doi:10.1016/j.prp.2019.152600
20. Wu JH, Tian XY, An QM, Guan XY, Hao CY. LINC00963 promotes hepatocellular carcinoma progression by activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22(6):1645–1652. doi:10.26355/eurrev_201803_14574
21. Peng T, Zhou L, Zuo L, Luan Y. MiR-506 functions as a tumor suppressor in glioma by targeting STAT3. Oncol Rep. 2016;35(2):1057–1064. doi:10.3892/or.2015.4406
22. Liang J, Liu N, Xin H. Knockdown long non-coding RNA PEG10 inhibits proliferation, migration and invasion of glioma cell line U251 by regulating miR-506. Gen Physiol Biophys. 2019;38(4):295–304. doi:10.4149/gpb_2019018
23. Sun G, Liu Y, Wang K, Xu Z. miR-506 regulates breast cancer cell metastasis by targeting IQGAP1. Int J Oncol. 2015;47(5):1963–1970. doi:10.3892/ijo.2015.3161
24. Li Z, Liu Z, Dong S, et al. miR-506 inhibits epithelial-to-mesenchymal transition and angiogenesis in gastric cancer. Am J Pathol. 2015;185(9):2412–2420. doi:10.1016/j.ajpath.2015.05.017
25. Song Z, Wang H, Zong F, Zhu C, Tao Y. MicroRNA-506 regulates apoptosis in retinoblastoma cells by targeting sirtuin 1. Cancer Manag Res. 2019;11:8419–8429. doi:10.2147/CMAR.S211122
26. Tan HY, Wang C, Liu G, Zhou X. Long noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer development through targeting STAT3. J Cell Biochem. 2019;120(4):4827–4836. doi:10.1002/jcb.26691
27. Yong W, Yu D, Jun Z, et al. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018;9(9):861. doi:10.1038/s41419-018-0908-z
28. Lei R, Xue M, Zhang L, Lin Z. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. Onco Targets Ther. 2016;10:35–46. doi:10.2147/OTT
29. Tönjes M, Barbus S, Park YJ, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19(7):901–908. doi:10.1038/nm.3217
30. Zheng YH, Hu WJ, Chen BC, et al. BCAT1, a key prognostic predictor of hepatocellular carcinoma, promotes cell proliferation and induces chemoresistance to cisplatin. Liver Int. 2016;36(12):1836–1847. doi:10.1111/liv.13178
31. Zhou W, Feng X, Ren C, et al. Over-expression of BCAT1, a c-Myc target gene, induces cell proliferation, migration and invasion in nasopharyngeal carcinoma. Mol Cancer. 2013;12:53. doi:10.1186/1476-4598-12-53
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.