Back to Journals » OncoTargets and Therapy » Volume 13
LINC00680 Promotes the Progression of Non-Small Cell Lung Cancer and Functions as a Sponge of miR-410-3p to Enhance HMGB1 Expression
Authors Wang H, Feng L , Zheng Y, Li W, Liu L, Xie S, Zhou Y, Chen C, Cheng D
Received 12 May 2020
Accepted for publication 28 July 2020
Published 18 August 2020 Volume 2020:13 Pages 8183—8196
DOI https://doi.org/10.2147/OTT.S259232
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Hui Wang,1,2 Li Feng,3 Yuqiong Zheng,2 Wen Li,2 Liang Liu,2 Sheng Xie,2 Yu Zhou,2 Chaofeng Chen,2 Deyun Cheng1
1Department of Respiratory and Critical Care Medicine, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, People’s Republic of China; 2Department of Respiratory and Critical Care Medicine, Chengdu First People’s Hospital, Chengdu, Sichuan 610051, People’s Republic of China; 3Department of Radiology, Chengdu First People’s Hospital, Chengdu, Sichuan 610051, People’s Republic of China
Correspondence: Deyun Cheng
Department of Respiratory and Critical Care Medicine, West China Hospital, Sichuan University, No. 37 Guoxue Alley, Wuhou District, Chengdu, Sichuan 610041, People’s Republic of China
Tel +86-28-85422114
Email [email protected]
Purpose: LINC00680 was reported to be involved in various cancers through multiple mechanisms. Therefore, we intended to investigate its role in the progression of non-small cell lung cancer (NSCLC).
Materials and Methods: Firstly, quantitative real-time polymerase chain reaction (qRT-PCR) was used to test LINC00680 in NSCLC tissue and cell lines. Subsequently, A549 and H1299 cells were transfected with LINC00680 overexpressing plasmids and their proliferation and colony formation and apoptosis was tested by Transwell assay and flow cytometry. In addition, xenograft tumor experiments in nude mice also affirmed. Meanwhile, we predicted that miR-410-3p, LINC00680 and high-mobility group protein box 1(HMGB1) relationship by Starbase, dual-luciferase reporter and RIP assay. Finally, the carcinogenic effects of LINC00680 were reversed by ethyl pyruvate (EP), a specific inhibitor of HMGB1.
Results: LINC00680 was upregulated in NSCLC and was closely related to the malignancy and poor prognosis of NSCLC patients. LINC00680 promoted proliferation and colony formation and inhibited apoptosis of A549 and H1299 cells. In addition, overexpressing LINC00680 accelerated the growth of NSCLC cells in xenograft tumor experiments in nude mice also affirmed. Meanwhile, high-mobility group protein box 1(HMGB1) was astoundingly amplified in NSCLC and was negatively regulated by miR-410-3p. Further, HMGB1 acted as a downstream target of miR-410-3p, upregulating miR-410-3p to attenuate HMGB1, while LINC00680 strengthened the expression of HMGB1 in A549 and H1299 cells.
Discussion: Thus, these results indicated that LINC00680 was cancerogenic in NSCLC by upregulating HMGB1 via sponging miR-410-3p.
Keywords: non-small cell lung cancer, LINC00680, miR-410-3p, HMGB1, progression
Introduction
Lung cancer is one of the malignancies with the highest fatality rate in China, among which non-small cell lung cancer (NSCLC) accounts for 80%~85%, and the 5-year survival rate is only 5%~10%.1 However, due to the lack of effective tumor molecular markers contributing to early clinical diagnosis and treatment, the therapeutic effect of NSCLC remains poor.2 Therefore, the exploration of new approaches to NSCLC treatment, especially the elucidation of NSCLC development mechanism at the gene level and the search for valid gene therapy targets, are hopefully to provide new theoretical basis and insight for its treatment.
LncRNAs are long non-coding RNAs without protein-coding functions and with more than 200 nucleotides in length. Recently, the levels of lncRNAs have been found to be altered in multiple cancers, and their roles in tumor regulation have attracted increasing attention. Similarly, the regulation of lncRNAs in NSCLC has been increasingly reported, while the roles and mechanisms of different lncRNAs are diversified. For example, Qu et al discovered that Linc-ROR was distinctly overexpressed in NSCLC tissues, which was an essential predictor of poor prognosis and played a carcinogenic role in NSCLC progression.3 Meanwhile, the functions of other lncRNAs, such as lncRNA BLACAT14 and linc-ITGB1,5 have also been reported in NSCLC. Moreover, previous studies have demonstrated that LINC00680 was an independent prognostic indicator for poor prognosis of the patients with soft tissue sarcoma,6 suggesting that LINC00680 exerts a role in tumorigenesis. Nevertheless, the function of LINC00680 in NSCLC was scarcely researched.
MiRNAs are a type of endogenous single-stranded non-coding RNAs with 20 to 24 nucleotides in length. MiRNAs can not only regulate cell development and differentiation, but also are potential tumor molecular markers involved in multiple cancer-related processes. In the past few years, the regulation of the lncRNA/miRNA axis in tumors has received growing attention. For example, lncRNA SNHG12 regulates the metastasis and growth of osteosarcoma by targeting the miR-195-5p/IGF1R axis.7 As a vital miRNA, miR-410-3p serves whether as an oncogene or tumor suppressor gene in different tumors.8 Nevertheless, the function of the lncRNA-mediated miR-410-3p in tumors remains largely unknown.
Studies have found that as a vital predictor of poor prognosis, HMGB1 is overexpressed in NSCLC,9 and reported to be regulated by a variety of miRNAs. For instance, it was discovered that miR-520a-3p weakens tumor cell growth by attenuating HMGB1, the Bcl-2/Bax ratio, and MMP-2 and MMP-9 expression, and enhancing Caspase-3.10 Moreover, miR-200c inhibits the EMT, invasion and migration of NSCLC cells by targeting HMGB1.11 Thus, HMGB1 functions as a carcinogenic factor in NSCLC.
Here, we explored the expression characteristics of LINC00680 in NSCLC and then conducted functional experiments of LINC00680 in NSCLC. Our data suggested that LINC00680 is upregulated in NSCLC tissues and predicts poorer survival of NSCLC patients. Functionally, LINC00680 upregulated HMGB1 via sponging miR-410-3p. Overall, this study illustrated a new biomarker and therapeutic target for lung cancer diagnosis and treatment.
Materials and Methods
Clinical Specimen Collection and Treatment
Tumor tissues and the matched adjacent normal tissues of 49 NSCLC patients (general information is shown in Table 1) who underwent surgical treatment in West China Hospital from October 2013 to October 2014 were chosen. No patients received any adjuvant treatment such as chemotherapy and radiotherapy before the surgery. The normal tissues were obtained from the para-cancerous tissues of the patients from the same period, and no cancer cells were observed through pathological examination. All specimens were stored in −196°C liquid nitrogen immediately after removal. This study was approved by the ethics committee of West China Hospital, and all of the patients concerned signed the informed consent. The follow-up period was from the date of surgery or pathological biopsy to December 20, 2019, and there was no missed case.
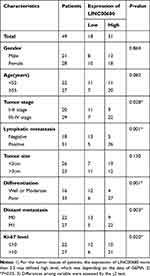 |
Table 1 Relationship Between LINC00680 Expression Level and Clinical Characteristics in NSCLC Tissue Samples |
Cell Culture
Normal bronchial epithelial cell lines (BEAS-2B) and NSCLC cell lines (A549, GLC82, H1299, H1975, NIC-H157 and SW1573) were purchased from ATCC (Rockville, USA). All of these cells were cultured in RPMI-1640 complete medium containing 10% fetal bovine serum (FBS) (Thermo Scientific Hyclone, Utah, USA) and 1% penicillin/streptomycin (Thermo Scientific Hyclone, Utah, USA), and incubated at 37°C with 5% CO2 and saturated humidity. The medium was exchanged every 2–3 days. The cells were trypsinized with 0.25% trypsin (Thermo Scientific Hyclone, Utah, USA) when reaching 90% confluence.
Cell Transfection
A549 and H1299 cells in the logarithmic growth phase were inoculated into 6-well plates at 5×106/well after trypsinization and passage. After the cell growth was stable, they were transfected with LINC00680 overexpressing plasmids and the matched negative controls, respectively following the instructions of the FuGENEHD Transfection Reagen (Roche, Shanghai, China). Subsequently, they were transfected with miR-410-3p mimics and the corresponding negative controls, respectively. Later on, the cells were cultured in an incubator at 37°C with 5% CO2. Twenty-four hours after the transfection, the total RNAs from cells were extracted for qRT-PCR to examine the expression alteration of LINC00680 and miR-410-3p in the transfected cells.
qRT-PCR
Total RNA was extracted from cells with the TRIzol reagent (Invitrogen, Shanghai, China), and then reverse-transcribed into cDNA with the PrimeScript ™ RT Reagent kit (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. PCR was performed using Bio-Rad CFX96 quantitative PCR system and SYBR following the manufacturer’s specifications as follows: pre-denaturation (95 °C, 5min), denaturation (95°C, 15s) and annealing (60°C, 30s). GAPDH served as internal contrast of LINC00680 and HMGB1, while U6 served as that of miR-410-3p. Meanwhile, the 2(-ΔΔCt) method was adopted for statistics. Primers used in this study were as follows: miR-410-3p: forward primer 5ʹ-AACCGGTAGCAGCACAGAAATG-3ʹ, reverse primer 5ʹ-CAGTGCAGGGTCCGAGGT-3ʹ. LINC00680: forward primer 5ʹ-CGATGTTTAAGCGTCCGGG-3ʹ, reverse primer 5ʹ-GGAAAGATGATGCGCTGTGT-3ʹ.
HMGB1: forward 5ʹ-GTGAACTGCTGCACGAAGAA-3ʹ, reverse primer 5ʹ-GCCTTTGAAATGTGCTCCCA-3. GAPDH: forward primer5ʹ-TGGTTGAGCACAGGGTACTT-3ʹ, reverse primer 5ʹ-CCAAGGAGTAAGACCCCTGG-3. U6: forward primer 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, reverse primer 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
CCK8 Assay
Forty-eight hours of transfection, A549 and H1299 cells in the logarithmic growth phase were trypsinized and adjusted to 2×103/mL, and then seeded in 96-well plates (100 μL of cell suspension per well), and three repetitive wells were set in each group. Afterward, the 96-well plates were placed in an incubator for further culture. Twenty-four hours later, 10 μL of CCK8 buffer (Biossci, Shanghai, China) was added to each well and cultured for 1 hour. After the culture was terminated, the 96-well plates were placed in an enzyme marker to quantify the optical density (OD value) of each well at the wavelength of 450 nm. The OD was then measured at 24, 48, 72 and 96 hours, respectively.
Colony Formation Experiment
Forty-eight hours of transfection, the cells in the logarithmic growth phase were inoculated into a 60 mm dish containing medium at 800 cells per dish. The culture medium was discarded 8 days later, and then the cells were rinsed with PBS three times. Subsequently, the cells were fixed with methanol for 20 min, stained with 1% methylene blue for 40 min, cleaned with deionized water twice and dried. Finally, the number of colonies (more than 50 cells) was calculated under a 40-fold microscope, and 3 repetitive wells were set for each group.
Flow Cytometry
Forty-eight hours of transfection, flow cytometry was conducted to analyze apoptosis. A549 and H1299 cells in the logarithmic phase were inoculated in 6-well plates, and got treated according to the experiment after being adherent the next day. Then, the cells were rinsed twice with pre-cooling PBS, and the cell number was adjusted at 1×106/mL in staining buffer. Afterward, 100 μL of cell suspension (1×105 cells) was added to the test tube, which was supplemented with 5 μL of PE Annexin-V and 7AAD each, and then cultured at room temperature in the darkness for 15 min. One hour later, flow cytometry was adopted to examine apoptosis in each group. The apoptosis rate was calculated as early apoptosis rate (LR) plus late apoptosis rate (UR). Each group had three parallel groups.
Transwell Assay
Forty-eight hours of transfection, A549 and H1299 cells were trypsinized with 0.25% trypsin, then they were centrifuged, resuspended, and dispersed in every single well of a 24-well culture plate. Chambers (8 µm poresize; Corning, Beijing, China) were coated with matrigel in the invasion experiment, while that of the migration experiment were not coated. Briefly, 5×104 transfected cells were added to the upper chamber, and the matrigel was supplemented. In contrast, the lower chamber was supplemented with a medium containing 10% FBS, and filled with 400 μL of RPMI-1640. After incubation at 37°C for 24 hours, the cells penetrated through the membranes were removed from the upper chambers. Subsequently, transwell membranes were fixed with 4% paraformaldehyde for 10 min and then stained with 0.5% crystal violet. After rinsed under running water, the cells were counted under an inverted microscope. All experiments were made in triplicate.
Dual-Luciferase Reporter Assay
All luciferase reporter vectors (LINC00680-WT, lLINC00680-MUT, HMGB11-WT and HMGB1-MUT) were designed by Promega (Promega, Madison, WI, USA). A549 cells (4.5×104) were inoculated in 48-well plates and cultured to 70% confluence. Then, LINC00680-WT, LINC00680-MUT, HMGB1-WT, and HMGB11-MUT were co-transfected with miR-410-3p mimics or negative controls into A549 cells using liposome 2000. Forty-eight hours after the transfection, the luciferase activity was determined according to the manufacturer’s instructions. All experiments were in triplicate.
Western Blot
A549 and H1299 cells were inoculated in 6-well plates (6×105/well) respectively, and got treated according to grouping. After 72 hours of culture, total protein was extracted, and the BCA method was adopted to assess the protein concentration. Then, proteins were isolated on 10% polyacrylamide gel electrophoresis and transferred to the membranes. Afterwards, the membranes were blocked with skimmed milk and incubated with primary antibodies of HMGB1 (ab3047, 1:1000, Abcam), Vimentin (ab3047, 1:1000, Abcam), N-cadherin (ab3047, 1:1000, Abcam), E-cadherin (ab22759, 1:1000, Abcam) and GAPDH (ab22759, 1:1000, Abcam) at 4°C overnight. Subsequently, the membranes were incubated with goat anti-rabbit secondary antibody at room temperature for 2 hours. Finally, ECL chemiluminescence reagent was adopted for color rendering, gel imaging system was employed for imaging, and ImageJ software was conducted for grayscale analysis, with GAPDH as the internal reference.
RIP Experiment
RIP assay was carried out with the Magna RipRNA and protein immunoprecipitation kit (Millipore, Bedford, MA, USA) according to the manufacturer’s protocol. Firstly, A549 and H1299 cells at 80% confluence were collected and lysed with Rip lysis buffer. Then, the anti-ago2 antibody (Abcam, Shanghai, China) was used for Ago2 immunoprecipitation, and immunoglobulin G (IgG) antibody served as the negative control. Finally, immunoprecipitated RNA was isolated, and the abundance of LINC00680 and HMGB1 in the binding part was observed by qRT-PCR.
Tumor Formation in Nude Mice
The A549 and H1299 cells overexpressing LINC00680 and the matched negative control cells were subcutaneously inoculated into the 6-week-old BALB/c-nu nude mice (2×107 cells per nude mouse) to construct the tumor formation model in vivo. The specific procedures were briefly described as follows: firstly, cells were trypsinized with 0.25% trypsin and harvested. Then, the cells were washed with serum-free medium and resuspended to get single-cell suspension (3×108/mL). Subsequently, 40 nude mice were divided into four groups (10 mice per group), and 0.1 mL of cell suspension was injected subcutaneously into the armpit of the left forelimb of each nude mouse. Finally, tumor diameter was measured at the first week, second week, 3rd week, fourth week and fifth week after the injection, and the mice were anesthetized five weeks later to determine the tumor size and weight, and the mice survival rate, weight and survival status were monitored. All animal experiments got the approval of the West China Hospital Animal Care and Use Committee.The experimental study was conducted per the National Institutes of Health Guide for the care and use of laboratory animals.
Data Analysis
SPSS20.0 statistical software (SPSS Inc., Chicago, IL, USA) was applied for data analysis. The measurement data were expressed as mean ± standard deviation (x±s). One-way ANOVA was adopted for the multi-factor comparison, and the t test was employed for the comparison between the two groups. P<0.05 was considered to be statistically significant.
Results
Expression of LINC00680 in NSCLC Tissues and Its Clinical Significance
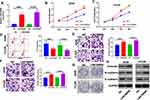
We adopted qRT-PCR experiments to verify the level of LINC00680 in NSCLC. The results showed that LINC00680 was distinctly upregulated in tumor tissues compared with that in normal adjacent tissues (P <0.05, Figure 1A). Meanwhile, through GEPIA (http://gepia.cancer-pku.cn), it was also manifested that LINC00680 was upregulated in Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), two types of NSCLC, as compared with that in the normal group (P <0.05, Figure 1B). Additionally, the LINC00680 expression in NSCLC tumor cell lines (A549, GLC82, H1299, H1975, NCI-H157, and SW1573) was notably higher than that in normal lung epithelial cells (P <0.05, Figure 1C). By analyzing the relationship between the clinical characteristics of NSCLC patients and the LINC00680 expression, we discovered that LINC00680 overexpression was correlated with higher tumor stage, worse differentiation, more obvious metastasis, and higher Ki67 index (Table 1). Further analysis of the patients’ survival confirmed that the higher LINC00680 expression predicted worse prognosis (P <0.05, Figure 1D). The above results suggested that LINC00680 may be a predictor of unfavorable prognosis in NSCLC as an oncogene.
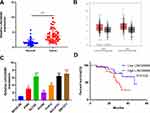 |
Figure 1 The LINC00680 expression in NSCLC tissues and cells. (A) qRT-PCR was conducted to examine the LINC00680 level in tumorous and adjacent normal tissues, ***P <0.001; (B) LINC00680 expression in LUAD and LUSC was analyzed through GEPIA (http://gepia.cancer-pku.cn). (C) qRT-PCR was employed to test the LINC00680 expression in normal bronchial epithelial cell lines (BEAS-2B) and NSCLC cell lines (A549, GLC82, H1299, H1975, NCI-H157, and SW1573), **P <0.01, ***P <0.01 vs BEAS-2B group. (D) Analysis of survival of patients with different LINC00680 levels. The expression of LINC00680 more than 2.5 was defined high level. |
Overexpressing LINC00680 Promoted the Growth and Metastasis of NSCLC Cells
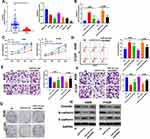
We constructed the LINC00680 overexpressing model in A549 and H1299 cells to further explore the role of LINC00680 in NSCLC (P <0.05, Figure 2A). Firstly, CCK8 and flow cytometry experiments affirmed that cell proliferation was enhanced, while the apoptosis was dramatically weakened after LINC00680 overexpression (compared with the NC group) (P <0.05, Figure 2B-D). Then, we adopted the transwell assay to investigate the LINC00680 function in cell migration and invasion. The results proved that overexpressing LINC00680 markedly enhanced the invasion and migration of A549 and H1299 cells (P <0.05, Figure 2E-F). Besides, the colony formation test testified that the cell colony formation was elevated by LINC00680 (Figure 2G). Further, we detected the epithelial-mesenchymal transition (EMT) changes in NSCLC cells via Western blot. The results indicated that Vimentin and N-cadherin were remarkably upregulated, while E-cadherin was downregulated after the LINC00680 overexpression (P <0.05, Figure 2H). The above results illustrated that LINC00680 functioned as an oncogene by accelerating the malignant biological behaviors of NSCLC cells.
Overexpressing LINC00680 Facilitated the Growth of NSCLC in vivo
For the purpose of verifying the function of LINC00680 in NSCLC progression in vivo, we conducted tumor formation experiments in nude mice with A549 and H1299 cells overexpressing LINC00680 (P<0.05, Figure 3A). It turned out that overexpressing LINC00680 elevated tumor size and weight (P <0.05, Figure 3B-D). Additionally, the result of Western blot suggested that Ki67, Vimentin, and N-cadherin were all distinctly upregulated, while E-cadherin was downregulated in tumors overexpressing LINC00680 (P <0.05, Figure 3E). These results further validated that LINC00680 aggravated the progression of NSCLC.
LINC00680 Functioned by Sponging miR-410-3p
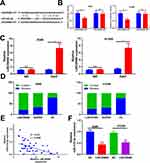
To further explore the downstream molecule of LINC00680, we employed the Starbase (http://starbase.sysu.edu.cn/) to analyze its potential target genes and discovered that miR-410-3p was the one (Figure 4A). Besides, the dual-luciferase reporter assay supported that miR-410-3p mimic obviously reduced the luciferase activity of LINC00680-WT, but had little effect on that of LINC00680-MT (P> 0.05, Figure 4B). In addition, the RIP experiment was used to testify the targeting relationship between LINC00680 and miR-410-3p. The results revealed that LINC00680 was more enriched in A549 and H1299 cells incubated with Ago2 than that of the IgG group (P <0.05, Figure 4C), suggesting that LINC00680 bound to Ago2 antibody through miR-410-3p mimic. Moreover, the localization of LINC00680 in the cytoplasm and nucleus of A549 and H1299 cells was detected by qRT-PCR. The results demonstrated that LINC00680 was mainly expressed in the cytoplasm, suggesting that LINC00680 sponged miRNAs as a competitive endogenous RNA (ceRNAs) in the cytoplasm (P> 0.05, Figure 4D). Besides, Pearson correlation coefficient revealed that LINC00680 was negatively related to the miR-410-3p expression in NSCLC tissues (r = −0.499, P <0.001, Figure 4E). Meanwhile, the miR-410-3p expression was distinctly dampened after LINC00680 overexpression (P <0.05, Figure 4F). These results indicated that LINC00680 bound to miR-410-3p, and exerted its functions by sponging miR-410-3p.
MiR-410-3p Attenuated the NSCLC Evolvement and Was Regulated by LINC00680
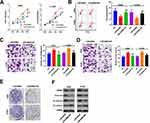
In consideration of the interaction between miR-410-3p and LINC00680 in NSCLC, we were also curious about the role of miR-410-3p in NSCLC. The results of qRT-PCR showed that miR-410-3p was downregulated both in NSCLC tissues (compared with the adjacent normal tissues) and cells (compared with the normal lung epithelial cells) (P < 0.05, Figure 5A). In addition, we conducted compensation experiments to explore the effects of LINC00680 and miR-410-3p in NSCLC development. It turned out that miR-410-3p expression was dramatically weakened after overexpressing LINC00680 (P <0.05, Figure 5B). Functionally, CCK8 and flow cytometry demonstrated that upregulating miR-410-3p weakened proliferation and enhanced apoptosis of A549 and H1299 cells, while the effects were obviously attenuated by overexpressing LINC00680 (P <0.05, Figure 5C-D). Further, A549 and H1299 cell migration and invasion were dampened after miR-410-3p overexpression compared with the control group, and they were further restrained after overexpressing LINC00680 (P <0.05, Figure 5E-F). Moreover, miR-410-3p mimics dampened colony formation as well as EMT of A549 and H1299 cells, while LINC00680 reversed the above effects (Figure 5G-H). These results indicated that miR-410-3p attenuated the progression of NSCLC and was inhibited by LINC00680.
HMGB1 Was a Target of miR-410-3p
By analyzing the potential target genes of miR-410-3p through Starbase (http://starbase.sysu.edu.cn/), we found that HMGB1 was a candidate target of it (Figure 6A). In addition, the dual-luciferase reporter assay and RIP assay revealed that miR-410-3p targeted the 3ʹUTR of HMGB1 (P> 0.05, Figure 6B and C). Further, the HMGB1 level in NSCLC tissues was examined, and it was proved to be notably increased compared with that in adjacent normal tissues (P <0.05, Figure 6D and E). Meanwhile, linear regression analysis indicated that HMGB1 was reversely correlated with the miR-410-3p expression, but positively correlated with the LINC00680 level (P <0.05, Figure 6F and G). Similarly, the expression of HMGB1 and LINC00680 was positively correlated in LUAD and LUSC as analyzed in GEPIA (http://gepia.cancer-pku.cn) (P <0.05, Supplement Figure 1 A and B). Furthermore, we detected the HMGB1 level after regulating miR-410-3p and LINC00680, and found that HMGB1 was overexpressed after augmenting LINC00680 but lowly expressed by miR-410-3p upregulation (Figure 6H). These data suggested that there was a regulatory axis of LINC00680-miR-410-3p-HMGB1 in NSCLC.
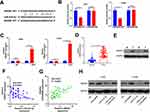 |
Figure 6 HMGB1 was the target of miR-410-3p. (A) The target genes of miR-410-3p were predicted by Starbase (http://starbase.sysu.edu.cn/) and HMGB1 had binding sites with miR-410-3p. (B) The-dual luciferase reporter assay was designed to reveal the relationship between HMGB1 and miR-410-3p; (C) RIP assay was performed to test the relationship between HMGB1 and miR-410-3p, and the level of HMGB1 was determined by qRT-PCR. D and E. the HMGB1 level in NSCLC tissues was examined by qRT-PCR (D) and Western blot (E). F and G. Linear regression analysis to test the correlations between HMGB1 and miR-410-3p (F), HMGB1 and LINC00680 (G). (H) Western blot was used to detect HMGB1 level after regulating miR-410-3p and LINC00680. N.S P>0.05, **P<0.01, ***P<0.001. |
Inhibiting HMGB1 Suppressed LINC00680-Mediated Effects on NSCLC
To further verify the role of HMGB1 in LINC00680-induced carcinogenic effects on NSCLC, the HMGB1 inhibitor EP was adopted to treat A549 and H1299 cells overexpressing LINC00680. The results showed that the EP administration reduced cell proliferation, enhanced apoptosis, and weakened cell migration, invasion and colony formation in NSCLC (compared with the LINC00680 group) (P <0.05, Figure 7A-E). Besides, the result of Western blot demonstrated that HMGB1, Vimentin, and N-cadherin were obviously downregulated, while E-cadherin was upregulated after the EP treatment (P <0.05, Figure 7F). These results illustrated that HMGB1 was a vital factor in LINC00680-induced accelerated progression of NSCLC.
Discussion
Due to the slow division and late metastasis of NSCLC, most of the patients are already in the middle or advanced stage once diagnosed, and the lack of biomarkers for early diagnosis has become one of the greatest challenges in its treatment.12 However, with the continuous exploration, increasing studies have confirmed that the lncRNA-miRNA axis affects the occurrence and progression of NSCLC. Here, our data indicated that there is a novel axis of LINC00680/miR-410-3p/HMGB1 in the progression of NSCLC (Figure 8).
Studies have shown that lncRNAs play important roles in the ceRNA network and regulate the expression of tumor-related genes, thus modulating the development of multiple tumors, whether by functioning as oncogenes or tumor suppressor genes.13 For example, LINC-PINT weakens the growth and colony formation of A549 and H1299, and inhibits the migration and invasion of NSCLC cells by regulating different genes.14,15 On the other side, studies have also suggested that lncRNAs aggravate NSCLC progression. For instance, Ye et al found that lncRNA-BLACAT1 accelerates the growth, migration and metastasis of NSCLC by sponging miR-144.4 Also, Guo et al discovered that LINC-ITGB1 promotes EMT and tumor stemness of NSCLC cells by targeting Snail.5 Moreover, lncRNAs such as LINC01561,16 PVT117 and LINC0052518 serve as oncogenes in NSCLC. Therefore, further exploration of the functions of different lncRNAs in NSCLC is of great significance for understanding its mechanism. According to the biological information and the results of this study, it was found that LINC00680 was distinctly upregulated in NSCLC, and was closely related to unfavorable prognosis. Disappointingly, the role and mechanism of LINC00680 in NSCLC remain elusive. Rong-Quan et al initially studied the effect of LINC00680, and their reports confirmed that LINC00680 is an individual prognostic indicator of poor prognosis in patients with sarcoma.6 Tang et al reported that LINC00680 promotes the accumulation of EGR3 in glioblastoma by sponging miR-320b, thereby enhancing the malignant biological behaviors of glioblastoma cells.19 Here, both in vitro and in vivo experiments confirmed that LINC00680 accelerated the progression of NSCLC by increasing the growth and metastasis, and inhibiting apoptosis of NSCLC cells, giving further evidence that LINC00680 is an oncogene in the tumor.
In addition to lncRNAs, miRNAs have also been a researching hotspot in oncology. In recent years, accumulating studies have shown that miRNAs regulate NSCLC by targeting oncogenes.20,21 For example, miR-532-3p,22 miR-15a,23 miR-125a24 and so on contribute to inhibiting NSCLC development. Besides, miR-410 has been found to exert multiple effects on tumorigenesis. For instance, Liu et al supported that the miR-410 overexpression enhances cell viability, reduces apoptosis, and exerts a carcinogenic function in colon cancer.25 Also, Chen et al discovered that miR-410 regulates the expression of the autophagy-related gene ATG16L1, thereby abating autophagy and enhancing the chemosensitivity in osteosarcoma.26 Studies in oral squamous cell carcinoma have shown that downregulating miR-410 promotes cancer cell growth and invasion by targeting Wnt-7b.27 Zhang et al reported that the miR-410-3p overexpression dampens the growth, colony formation, and number of EdU-positive cells in MDA-MB-231 cells, and miR-410-3p attenuates EMT of breast cancer by targeting Snail.8 In this study, it was indicated that miR-410-3p was downregulated in NSCLC tissues and cells, while overexpressing miR-410-3p markedly weakened the proliferation, migration and invasion, and promoted apoptosis of NSCLC cells, suggesting that miR-410-3p suppresses NSCLC.
Current researches have confirmed that lncRNAs function as ceRNAs and regulate downstream genes by sponging miRNAs. For example, Geo et al reported that lncRNA LINC01561 aggravates the progression of NSCLC by regulating the miRNA-526b/EZH2 axis.16 Also, Qiu et al reported that lncRNA PVT1 plays a carcinogenic role by regulating miRNA-526b/EZH2.17 Our study confirmed that miR-410-3p was a target of LINC00680, which mainly exerted its function by sponging miR-410-3p in the cytoplasm. In addition, overexpressing LINC00680 notably inhibited the miR-410-3p expression in NSCLC and also crippled the tumor inhibitory effects of miR-410-3p. These results further verified that LINC00680 promotes cell growth and metastasis, while inhibits apoptosis in NSCLC via regulating miR-410-3p.
HMGB1 is a small DNA-binding protein involved in a variety of cellular processes, which is translocated from the nucleus to the cytoplasm through active or passive release. Evidence suggests that extracellular HMGB1 promotes tumor growth by enhancing inflammation, EMT, cell migration, and angiogenesis. Recent studies have indicated that HMGB1 influences cancer development by interacting with miRNAs.28 For example, Meng et al reported that Rh-Endostatin induces the HMGB1 inhibition in multiple NSCLC cells dose- and time-dependently, thus attenuating A549 cell proliferation.29 Besides, the relationship between miRNA expression and HMGB1 has been affirmed in various diseases, including NSCLC. Some scholars have reported that miR-520a-3p notably reduces the HMGB1 mRNA and protein levels, inhibits proliferation, migration and invasion of NCI-H157 cells, and induces apoptosis.10 Moreover, miR-200C,11 miR-142-3p,30 and microRNA-181b31 have been reported to slow the progression of NSCLC by regulating HMGB1. Interestingly, it was found that in pancreatic cancer that miR-410-3p enhances the chemosensitivity to gemcitabine by inhibiting the HMGB1-induced PDAC cell autophagy during chemotherapy.32 However, it remains unclear whether miR-410-3p dampens NSCLC progression by regulating HMGB1. The results of this study confirmed that the inhibition of HMGB1 weakens the malignant biological behaviors of NSCLC, while LINC00680 elevates HMGB1 and EMT of A549 and H1299 cells by sponging miR-410-3p, thus inhibiting growth, migration and invasion of NSCLC.
In conclusion, our study confirmed that LINC00680 is overexpressed in NSCLC tissues and cells, and overexpressing LINC00680 promotes the malignant biological behavior of NSCLC, elucidating the role and mechanism of the LINC00680/miR-410-3p/HMGB1 axis in the development of NSCLC. LINC00680, as an oncogene, elevates the HMGB1 expression by sponging miR-410-3p, thus affecting the proliferation, metastasis and apoptosis of cancer cells (Figure 8). This study better explored the molecular mechanism of LINC00680 in regulating the occurrence and development of NSCLC, which is conducive to its early diagnosis and treatment, and provides favorable theoretical support for the treatment and prognosis of NSCLC. However, more mechanisms need to be further studied.
Data Sharing Statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Statement
Our study was approved by the Ethics Review Board of West China Hospital, Sichuan University.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wakelee H, Kelly K, Edelman MJ. 50 years of progress in the systemic therapy of non-small cell lung cancer. Am Soc Clin Oncol Educ Book. 2014;177–189. doi:10.14694/EdBook_AM.2014.34.177
2. Brinkmeyer JK, Moore DC. Necitumumab for the treatment of squamous cell non-small cell lung cancer. Oncol Pharm Pract. 2016;2016.
3. Qu C-H, Sun Q-Y, Zhang F-M, et al. Long non-coding RNA ROR is a novel prognosis factor associated with non-small-cell lung cancer progression. Eur Rev Med Pharmacol Sci. 2017;21:4087–4091.
4. Ye J-R, Liang L, Feng Z. Long non-coding RNA bladder cancer associated transcript 1 promotes the proliferation, migration, and invasion of nonsmall cell lung cancer through sponging miR-144. DNA Cell Biol. 2017;36(10):845–852. doi:10.1089/dna.2017.3854
5. Guo L, Cencen S, Shilei X, et al. Knockdown of long non-coding RNA linc-ITGB1 inhibits cancer stemness and epithelial-mesenchymal transition by reducing the expression of Snail in non-small cell lung cancer. Thorac Cancer. 2019;10(2):128–136. doi:10.1111/1759-7714.12911
6. Rong-Quan H, Qing-Jun W, Rui-Xue T, et al. Prediction of clinical outcome and survival in soft-tissue sarcoma using a ten-lncRNA signature. Oncotarget. 2017;8:80336–80347. doi:10.18632/oncotarget.18165
7. Ning X, Jiayuan X, Zhuan Z, et al. Downregulation of lncRNA SNHG12 reversed IGF1R-induced osteosarcoma metastasis and proliferation by targeting miR-195-5p. Gene. 2020;726:144145. doi:10.1016/j.gene.2019.144145
8. Zhang Y-F, Yue Y, Wang-Zhao S, et al. miR-410-3p suppresses breast cancer progression by targeting Snail. Oncol Rep. 2016;36:480–486. doi:10.3892/or.2016.4828
9. Yi-Liang W, Ming-Hsien C, Ying-Erh C, et al. Association of EGFR mutations and HMGB1 genetic polymorphisms in lung adenocarcinoma patients. J Cancer. 2019;10(13):2907–2914. doi:10.7150/jca.31125
10. Lv X, Yao L, Nie Y-Q, et al. MicroRNA-520a-3p suppresses non-small-cell lung carcinoma by inhibition of High Mobility Group Box 1 (HMGB1). Eur Rev Med Pharmacol Sci. 2018;22:1700–1708. doi:10.26355/eurrev_201803_14583
11. Po-Len L, Wei-Lun L, Jia-Ming C, et al. MicroRNA-200c inhibits epithelial-mesenchymal transition, invasion, and migration of lung cancer by targeting HMGB1. PLoS One. 2017;12:e0180844. doi:10.1371/journal.pone.0180844
12. Kocher F, Pircher A, Mohn-Staudner A, et al. Multicenter phase III study evaluating docetaxel and cisplatin as neo-adjuvant induction regim en prior to surgery or radiochemotherapy with docetaxel,followed by adjuvant docetaxe[therapy in chemo naive patients with NSCLC stage 1I,III A and 11 I B (TAX-ATI.203Tria1). Lung Cancer. 2014;85(3):395–400. doi:10.1016/j.lungcan.2014.06.019
13. Yue W, Zihui F, Mei H, et al. Long-non-coding RNAs (lncRNAs) in drug metabolism and disposition, implications in cancer chemo-resistance. Acta Pharm Sin B. 2020;10(1):105–112. doi:10.1016/j.apsb.2019.09.011
14. Shu W, Wenyang J, Xinghua Z, et al. LINC-PINT alleviates lung cancer progression via sponging miR-543 and inducing PTEN. Cancer Med. 2020. doi:10.1002/cam4.2822
15. Libin Z, Jing H, Jiagui L, et al. Long non-coding RNA LINC-PINT inhibits non-small cell lung cancer progression through sponging miR-218-5p/PDCD4. Artif Cells Nanomed Biotechnol. 2019;47:1595–1602. doi:10.1080/21691401.2019.1605371
16. Geo W, Chao-Qun Q, Mao-Guo F, et al. SOX2-induced upregulation of lncRNA LINC01561 promotes non-small-cell lung carcinoma progression by sponging miR-760 to modulate SHCBP1 expression. J Cell Physiol. 2020.
17. Qiu C, Sai L, Datong S, et al. lncRNA PVT1 accelerates progression of non-small cell lung cancer via targeting miRNA-526b/EZH2 regulatory loop. Oncol Lett. 2020;19:1267–1272. doi:10.3892/ol.2019.11237
18. Zhiguang Y, Xingyu L, Peng Z, et al. Long non-coding RNA LINC00525 promotes the non-small cell lung cancer progression by targeting miR-338-3p/IRS2 axis. Biomed Pharmacother. 2020;124:109858. doi:10.1016/j.biopha.2020.109858
19. Wei T, Wang D, Lianqi S, et al. LINC00680 and TTN-AS1 stabilized by EIF4A3 promoted malignant biological behaviors of glioblastoma cells. Mol Ther Nucleic Acids. 2019;19:905–921. doi:10.1016/j.omtn.2019.10.043
20. Weidle Ulrich H, Fabian B, Adam N. MicroRNAs as potential targets for therapeutic intervention with metastasis of non-small cell lung cancer. Cancer Genomics Proteomics. 2019;16:99–119. doi:10.21873/cgp.20116
21. Ha KD, Sojung P, HyeongRyul K, et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1.4. Cancer Lett. 2020.
22. Wenhua J, Liangda Z, Qingqing Y, et al. MiR-532-3p inhibits metastasis and proliferation of non-small cell lung cancer by targeting FOXP3. J BUON. 2019;24:2287–2293.
23. Shuai G, Ming L, Juan L, et al. Inhibition mechanism of lung cancer cell metastasis through targeted regulation of Smad3 by miR-15a. Oncol Lett. 2020;19:1516–1522. doi:10.3892/ol.2019.11194
24. Huang H, Jingyu H, Jie Y, et al. miR-125a regulates HAS1 and inhibits the proliferation, invasion and metastasis by targeting STAT3 in non-small cell lung cancer cells. J Cell Biochem. 2020;121(5–6):3197–3207. doi:10.1002/jcb.29586
25. Liu C, Zhang A, Lei C, et al. miR‑410 regulates apoptosis by targeting Bak1 in human colorectal cancer cells. Mol Med Rep. 2016;14:467–473. doi:10.3892/mmr.2016.5271
26. Chen R, ZHOU X, He B, et al. MicroRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma. Mol Med Rep. 2017;15:1326–1334. doi:10.3892/mmr.2017.6149
27. Shine-Gwo S, Jenn-Ren H, Wei-Min C, et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 2014;74:7560–7572. doi:10.1158/0008-5472.CAN-14-0978
28. Qun G, Shumin W, Xinfeng C, et al. Cancer-cell-secreted CXCL11 promoted CD8 T cells infiltration through docetaxel-induced-release of HMGB1 in NSCLC. J Immunother Cancer. 2019;7:42. doi:10.1186/s40425-019-0511-6
29. Meng F-J, Shuo W, Yi-Jie Y, et al. Recombined humanized endostatin-induced suppression of HMGB1 expression inhibits proliferation of NSCLC cancer cells. Thorac Cancer. 2019;10:90–95. doi:10.1111/1759-7714.12905
30. Yuqing C, Xin Z, Jianou Q, et al. MiR-142-3p overexpression increases chemo-sensitivity of NSCLC by Inhibiting HMGB1-mediated autophagy. Cell Physiol Biochem. 2017;41(4):1370–1382. doi:10.1159/000467896
31. Yun L, Hu X, Daokui X, et al. MicroRNA-181b is downregulated in non-small cell lung cancer and inhibits cell motility by directly targeting HMGB1. Oncol Lett. 2016;12(5):4181–4186. doi:10.3892/ol.2016.5198
32. Junjie X, Dan W, Ailin W, et al. MicroRNA-410-3p attenuates gemcitabine resistance in pancreatic ductal adenocarcinoma by inhibiting HMGB1-mediated autophagy. Oncotarget. 2017;8(64):107500–107512. doi:10.18632/oncotarget.22494
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.