Back to Journals » Cancer Management and Research » Volume 13
LINC00665 Stimulates Breast Cancer Progression via Regulating miR-551b-5p
Authors Qi L, Sun B, Yang B, Lu S
Received 1 August 2020
Accepted for publication 18 November 2020
Published 5 February 2021 Volume 2021:13 Pages 1113—1121
DOI https://doi.org/10.2147/CMAR.S275096
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Liqiang Qi,1 Bo Sun,2 Beibei Yang,2 Su Lu2
1Department of Breast Surgical Oncology, Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100021, People’s Republic of China; 2The 2nd Department of Breast Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin, 300060, People’s Republic of China
Correspondence: Liqiang Qi
Department of Breast Surgical Oncology, Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, No. 17 Panjiayuan Nanli, Chaoyang District, Beijing, 100021, People’s Republic of China
Tel +86 10-87787130
Email [email protected]
Introduction: Long intergenic non-protein coding RNA 665 (LINC00665) has been revealed to contribute cancer progression in many cancer types including liver and gastric cancer. However, the roles of LINC00665 in breast cancer (BC) remain to be explored.
Methods: We explored LINC00665 expression in BC tissues and normal tissues at GEPIA. Then, its expression in BC cells (HCC-1937, MDA-MB-231, and MCF-7) and normal cells (MCF10A) was analyzed with qRT-PCR. In addition, the mechanisms of LINC00665 in BC were explored using bioinformatic analyses, luciferase activity reporter assay, RNA pull-down assay, and rescue experiments.
Results: We showed LINC00665 expression was significantly increased in both BC tissues and cells. The knockdown of LINC00625 significantly inhibits BC cell growth and promotes cell apoptosis in vitro, while the overexpression of LINC00625 has the opposite effects on BC progression. LINC00665 could affect BC progression via regulating miR-551b-5p.
Discussion: Taken together, our study showed that the LINC00665/miR-551b-5p axis was involved in the progression of BC.
Keywords: LINC00665, miR-551b-5p, breast cancer, progression
Introduction
Breast cancer (BC) is the major health threat for women worldwide and with an increasing trend.1 The prognosis of BC patients varies depending on the cancer diagnosis stage and molecular subtype.2 In the past few decades our understanding of the pathogenesis of BC has greatly improved but the prognosis is still not optimistic.3,4 Therefore, a deeper understanding of the mechanisms behind BC progression is still needed.
Mounting evidence has pointed out the importance of non-coding RNAs (ncRNAs) such as circulating RNA (circRNA), long non-coding RNA (lncRNA), and microRNA (miRNA) in BC.5–7 lncRNAs are genomic transcripts at the length of over 200 nucleotides with limited protein coding capacity but which play crucial roles in normal cellular functions maintenance and cell malignant transformation.8 For instance, growth arrest‑specific 5 (GAS5) is a tumor suppressive lncRNA and suppresses BC progression.6 LINC00160 may affect the response of BC cells to chemical reagents including paclitaxel and doxorubicin by regulating TFF3 expression.9 LncRNA DLX6-AS1 could regulate the epithelial-mesenchymal transition process of BC cells and their sensitivity to cisplatin via regulating miR-199b-5p.10
Long intergenic non-protein coding RNA 665 (LINC00665) is reported to stimulate cancer progression. Increased LINC00665 expression could promote gastric cancer tumorigenesis and can be used as a predictor for late TNM stage and poorer prognosis.11 Moreover, LINC00665 was found to accelerate the malignancy behaviors of hepatocellular carcinoma.12 In BC, LINC00665 plays an oncogenic role in affecting epithelial-mesenchymal transition-like phenotype via regulating the miR-379-5p/LIN28 axis.13 Although the carcinogenesis role of LINC00665 has been reported in BC, its detailed mechanism in BC remains to be explored.
We aimed to investigate the mechanisms of LINC00665 in BC. The impact of LINC00665 on BC cellular processes was analyzed using loss-of-function and gain-of-function experiments. Moreover, in vivo experiments were conducted to analyze the impact of LINC00665 on tumor growth.
Materials and Methods
Patient Samples
36 pairs of BC tumor tissues and adjacent tissues were obtained from the Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. The study protocol was approved by the ethic committee of the Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. Tissues were treated with RNAprotect reagent (Qiagen, Germany), frozen using liquid nitrogen and stored at −80 °C until usage. Informed written consent was obtained from all patients.
Cell Lines
Normal breast epithelial cell line (MCF10A) and BC cells (HCC-1937, MDA-MB-231, and MCF-7) were purchased from ATCC (Manassas, VA, USA). DMEM supplemented with 10% FBS (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) was used to incubate BC cells, while MEGM supplemented with cholera toxin was used to incubate MCF10A cells. All these cells were kept at the condition of 37 °C and 5% CO2.
Cell Transfection
Specific siRNA against LINC00665 (si-LINC00665) and negative control (si-NC), as well as miR-551b-5p mimic, miR-551b-5p inhibitor and negative control (mi-NC) were all available RiboBio (Guangzhou, Guangdong, China). Sequence of LINC00665 was inserted into pcDNA3.1 to overexpress LINC00665 by RiboBio. Cells incubated in a 24-well plate were subjected to transfection of these molecules in the presence of Lipofectamine 2000 (Invitrogen). Cells were collected for following analyses after 48 h transfection.
RNA Isolation and RT-qPCR Analysis
Tissues and cells were treated with a Trizol reagent (Invitrogen) to isolate RNA samples according to the provided protocols. cDNA was synthesized from RNA samples using a Reverse Transcription kit (Takara, Dalian, Liaoning, China). SYBR Green (Takara) and ABI 7900HT (Applied Biosystems, Foster City, CA, USA) were used to quantify gene expression levels with GAPDH and U6 snRNA as internal controls for LINC00665 and miR-551b-5p, respectively. Primers were: LINC00665: 5ʹ-GGTGCAAAGTGGGAAGTGTG-3ʹ (forward), 5ʹ-CGGTGGACGGATGAGAAACG-3ʹ; GAPDH: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ (forward), 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ (reverse); miR-551b-5p: 5ʹ-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGTCTC-3ʹ (forward), 5ʹ-GCAGGGTCCGAGGTATTC-3ʹ (reverse); U6 snRNA: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (forward), 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (reverse). Relative gene expression level was calculated with the 2−ΔΔCt method.
MTT Assay
Cell proliferation rate was measured with (3-(4,5-dimethyl-thiazol-2-yl) 2,5-diphenyl tetrazolium bromide) (MTT) assay. Cells were collected and seeded in a 96-well plate at the density of 4 × 103 cells/well. After 24, 48, 72 h transfection, MTT solution was added to each well and incubation was done for another 4 h. Then, the medium was discarded before the addition of DMSO reagent to dissolve formazan crystals. Optical density was measured with a microplate reader at the wavelength of 490 nm.
Colony Formation Assay
1000 cells were grown in the above-described medium and all were grown for 14 days. The visible colonies were fixed with ethanol, and stained with crystal violet. Numbers of colonies were counted using Image J software.
Cell Apoptosis Analysis
1 × 105 cells were collected and digested using trypsin. Then, cells were double stained with Annexin V and PI (Beyotime, Haimen, Jiangsu, China) in darkness for 15 min. Then, apoptosis rate was measured using flow cytometry (BD Biosciences, San Jose, CA, USA). Both the early stage and late stage apoptosis cells were counted as apoptosis cells.
Luciferase Activity Assay
DIANA LncBase Predicted V.2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted) was used to detect the interactions of lncRNA and miRNA. LINC00665 covered the wild-type or mutant miR-551b-5p binding sites were synthesized by RiboBio and inserted into pmirGLO dual-luciferase vector (Promega, Madison, WI, USA) to generate LINC00665-WT/MT. The above luciferase constructs were co-transfected with miR-551b-5p mimic or NC-mimic into BC cells using Lipofectamine 2000 for 48 h. Finally, relative luciferase activity was measured using a dual-luciferase activity reporter system (Promega).
RNA Pull-Down Assay
A RNA pull-down assay was employed to investigate the connection of LINC00665 and miR-551b-5p. The biotin labeled miR-551b-5p-WT, miR-551b-5p-MT, or mi-NC were synthesized by RiboBio. Cells which were transfected with these three molecules, respectively. 48 h later, the cells were lysated with RIPA lysis buffer and incubated with streptavidin conjugated with magnetic beads. Finally, the RNA of the pellets was isolated with Trizol reagent and subjected to RT-qPCR analysis.
Analysis of LINC00665 and miR-551b-5p in BC Tissues
Expression levels of LINC00665 in BC tissues and normal tissues were detected at GEPIA and ENCORI. In addition, the levels of miR-551b-5p in tissues were analyzed at UALCAN.
In vivo Tumor Growth Model
SPF grade 4-week-old BALB/c nude mice (n = 6 for each group, Shi Laike Company, Shanghai, China) were injected with LINC00665 stable knockdown MCF-7 cells at the right flank. Stable LINC00665 knockdown cells were generated by transfecting sh-LINC00665 containing pGPH1/Neo plasmid into cells, treated with antibiotic and isolating a single clone to obtain the stable transfected cells. Successful transfection was validated by RT-qPCR. The cells with sh-NC transfection were injected into mice and used as control. After injection, tumor volume was recorded every 7 days for a total of 4 times. 28 days later, mice were sacrificed to weigh the tumor tissues. Animal study protocol was approved by the animal ethic committee of the Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College and performed in accordance with the National Laboratory Animal Welfare ethics.
Statistical Analysis
Experiments were repeated for three times. Data analysis was conducted using GraphPad Prism 6.0 (La Jolla, CA, USA) and expressed as mean ± SD. ANOVA and Tukey post hoc tests were used to analyze differences among multiple groups, while Student’s t-test was used for comparison in two groups. Correlation between LINC00665 and miR-551b-5p was analyzed using the Person test. The survival curve was drawn using the Kaplan-Meier method, and the difference was analyzed using log rank test. A P value less than 0.05 was believed to indicate significant statistical differences.
Results
LINC00665 Expression Was Expression in BC
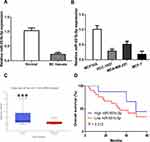
LINC00665 expression was a significantly upregulated expression in BC tissues compared with normal tissues (Figure 1A, P < 0.001). GEPIA and ENCORI analyses of LINC00665 in large numbers of BC patients showed LINC00665 had an increased expression in BC tissues compared with normal tissues (Figure 1B and C, P < 0.05). To explore the correlation of LINC00665 expression and the prognosis of BC patients, a Kaplan-Meier curve was conducted and we found high LINC00665 could predict poor overall survival of BC patients (Figure 1D, P = 0.016). In addition, we found high LINC00665 levels were correlated with TNM stage and lymph node metastasis of BC patients enrolled in this study (Table 1). Furthermore, we showed LINC00665 could be a possible biomarker to predict the overall survival of BC patients using univariate and multivariate analyses (Table 2).
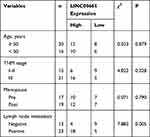 |
Table 1 Correlations of LINC00665 Expression and Clinicopathological Features of Breast Cancer Patients |
 |
Table 2 Univariate and Multivariate Analyses of Overall Survival in Patients with BC |
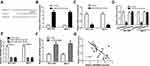
Silencing of LINC00665 Suppresses BC Cell Growth
LINC00665 expression was extremely high in BC cells relative to MCF-10A cells, especially the HCC-1937 and MCF-7 cells (Figure 2A, P < 0.001). Then, loss-of-function experiments were conducted to investigate the influence of LINC00665 on cell growth. The introduction of si-LINC00665 significantly decreased LINC00665 expression in HCC-1937 and MCF-7 cells (Figure 2B, P < 0.001). MTT assay showed cell proliferation ability declined after the silencing of LINC00665 (Figure 2C, P < 0.001). We also showed knockdown of LINC00665 inhibits colony formation (Figure 2D, P < 0.01). By contrast, cell apoptosis percentage (Q2+Q4 quadrants) was elevated after the knockdown of LINC00665 (Figure 2E, P < 0.01).
In addition, gain-of experiments were also conducted in this work. LINC00665 levels were revealed to be significantly elevated by pLINC00665 (Supplementary Figure 1A, P < 0.001). Subsequently, we found that forcing the expression of LINC00665 could augment cell proliferation (P < 0.001) by regulating colony formation (P < 0.01) and cell apoptosis (P < 0.01) (Supplementary Figure 1B-D).
LINC00665 Served as miR-551b-5p Sponge in BC
Through DIANA LncBase Predicted V.2 analysis, miR-551b-5p was found as a potential target of LINC00665 (Figure 3A, P < 0.001). miR-551b-5p mimic transfection increased, while miR-551b-5p inhibitor transfection decreased miR-551b-5p expression levels in BC cells (Figure 3B and C, P < 0.001). A luciferase activity reporter assay showed miR-551b-5p overexpression could decrease the relative luciferase activity of BC cells transfected with LINC00665-WT (P < 0.001), but not those with LINC00665-MT (Figure 3D). An RNA pull-down assay showed LINC00665 was enriched in the complexes pulled down by bio-miR-551-5p-WT compared with other groups (Figure 3E, P < 0.001). Moreover, we found miR-551b-5p expression level was increased by si-LINC00665 in BC cells (Figure 3F, P < 0.01). Thereafter, we found LINC00665 and miR-551b-5p was negatively correlated in BC tissues (Figure 3G).
miR-551b-5p Expression Was Decreased in BC
Then, we detected the importance of miR-551b-5p in BC. miR-551b-5p expression was decreased in BC tissues (P < 0.001) and cells (P < 0.001 in HCC-1937 and MCF-7 cells; P < 0.01 in MDA-MB-231 cells) (Figure 4A-C). Importantly, low miR-551b-5p level was an indicator for poorer overall survival of BC patients. (Figure 4D, P = 0.012)
LINC00665 Regulated BC Cell Growth via miR-551b-5p
To investigate the function of the LINC00665/miR-551b-5p axis in BC, we carried out rescue experiments. We validated the stimulation effect of si-LINC00665 on miR-551b-5p expression which could be abolished by miR-551b-5p inhibitor (Figure 5A). Expectedly, CCK-8 assay, colony formation and flow cytometry assay validated that the stimulation effects of si-LINC00665 on BC cell growth were counteracted by miR-551b-5p inhibitor (Figure 5B-D).
Role of LINC00665 on BC Tumor Growth in vivo
To determine the importance of LINC00665/miR-551b-5p axis in BC, we detected their effects in animal model. As expected, we discovered that LINC00665 knockdown hindered tumor growth in vivo (Figure 6A and B, P < 0.001). Analysis of the expression of LINC00665 and miR-551b-5p in LINC00665 knockdown tissues verified the downregulation status of LINC00665 and the upregulation status of miR-551b-5p (Figure 6C, P < 0.001).
Discussion
For a long period, lncRNAs were considered as noise in genomes, without biological roles.14 Increasingly evidence in recent years have indicated that lncRNAs are associated with tumor progression and drug resistance.6,9,10 Hence, studying lncRNA is useful to understand the mechanisms relevant to BC progression. This work focused on the regulatory mechanism of LINC00665 in BC with the aim to provide novel insights into the pathogenesis of BC.
Previous studies have shown LINC00665 has an oncogenic role in BC.13,15 For instance, Ji et al pointed out the LINC00665/miR-379-5p/LIN28B regulatory network influences the tumorigenesis of BC.13 Zhou et al pointed out that knocking down the LINC00665 expression could inhibit BC metastasis via regulating the epithelial-mesenchymal transition processes.15 In this study, we found LINC00665 expression was increased in BC tissues and cells. We also showed high LINC00665 levels are a predictor for the poorer overall survival of BC patients. These results manifested that LINC00665 expressed at a high level, indicating it might be effective in the progression of BC.
For the sake of exploring the functions of LINC00665 in BC, we performed in vitro and in vivo experiments. Our results showed that silencing LINC00665 suppresses BC cell proliferation by stimulating cell apoptosis in vitro. On the contrary, the augment of LINC00665 expression has the opposite effects on BC cell behaviors as compared with the knockdown of LINC00665. Previous studies have indicated that LINC00665 could stimulate cancer cell proliferation, migration, invasion.11–13 Hence, our results presented here are in accordance with these previous results that LINC00665 could promote cell growth. The limitation of this work is that we analyzed the effects of LINC00665 on the malignant behaviors of HCC-1937 and MCF-7, which belong to different subtypes of BC, but did not further analyze which LINC00665 has the most significant effects on which subtype of BC by analyzing more cells in vitro and also in vivo. Moreover, our in vivo analyses results using animal model also indicated that knockdown of LINC00665 hinders BC tumor growth. Taken together, our results implied the importance of LINC00665 in regulating BC progression.
miRNAs are single strand RNA the length of 18–24 nucleotides and can function as the downstream targets of lncRNAs.16 For example, miR-149-3p and miR-379-5p have been identified as the targets of LINC00665 in cancers.11,13 Jiang et al revealed that miR-551b-5p was strongly differentially expressed in gastric cancer and can efficiently distinguish healthy controls from gastric cancer patients.17 This time, miR-551b-5p was found to be a significantly decreased expression in BC and correlated with a worse overall survival of cancer patients. We also found that knockdown of miR-551b-5p significantly stimulated BC cell growth, and partially abolished the effects of LINC00665 knockdown on BC malignant behaviors. Although we provided evidence for the roles of the LINC00665/miR-551b-5p axis in BC, there is still room for improvement. First of all, the effects of LINC00665 on BC cell behaviors were not fully investigated. What is more, the downstream molecules or pathways regulated by LINC00665/miR-551b-5p were not investigated.
Conclusion
In conclusion, we propose the LINC00665/miR-551b-5p regulatory network that can mediate the progression of BC, which may be used as a target for cancer treatment.
Acknowledgment
This work was supported by the Natural Science Foundation of China (Grant No. 81001187).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomark Prev. 2017;26(4):444–457. doi:10.1158/1055-9965.EPI-16-0858
2. Mariotto AB, Etzioni R, Hurlbert M, Penberthy L, Mayer M. Estimation of the number of women living with metastatic breast cancer in the United States. Cancer Epidemiol Biomarkers Prev. 2017;26(6):809–815. doi:10.1158/1055-9965.EPI-16-0889
3. Martos T, Casadevall D, Albanell J. Circulating tumor cells: applications for early breast cancer. Adv Exp Med Biol. 2020;1220:135–146.
4. Mei J, Hao L, Wang H, et al. Systematic characterization of non-coding RNAs in triple-negative breast cancer. Cell Prolif. 2020;53(5):e12801. doi:10.1111/cpr.12801
5. Zhang L, Dong X, Yan B, Yu W, Shan L. CircAGFG1 drives metastasis and stemness in colorectal cancer by modulating YY1/CTNNB1. Cell Death Dis. 2020;11(7):542. doi:10.1038/s41419-020-2707-6
6. Li G, Qian L, Tang X, Chen Y, Zhao Z, Zhang C. Long non‑coding RNA growth arrest‑specific 5 (GAS5) acts as a tumor suppressor by promoting autophagy in breast cancer. Mol Med Rep. 2020;22(3):2460–2468. doi:10.3892/mmr.2020.11334
7. Guo F, Zhu X, Zhao Q, Huang Q. miR‑589‑3p sponged by the lncRNA TINCR inhibits the proliferation, migration and invasion and promotes the apoptosis of breast cancer cells by suppressing the Akt pathway via IGF1R. Int J Mol Med. 2020;46(3):989–1002. doi:10.3892/ijmm.2020.4666
8. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606
9. Wu H, Gu J, Zhou D, et al. LINC00160 mediated paclitaxel-And doxorubicin-resistance in breast cancer cells by regulating TFF3 via transcription factor C/EBPβ. J Cell Mol Med. 2020. doi:10.1111/jcmm.15487
10. Du C, Wang Y, Zhang Y, Zhang J, Zhang L, Li J. LncRNA DLX6-AS1 contributes to epithelial-mesenchymal transition and cisplatin resistance in triple-negative breast cancer via modulating Mir-199b-5p/Paxillin axis. Cell Transplant. 2020;29:963689720929983. doi:10.1177/0963689720929983
11. Qi H, Xiao Z, Wang Y. Long non-coding RNA LINC00665 gastric cancer tumorigenesis by regulation miR-149-3p/RNF2 axis. Onco Targets Ther. 2019;12:6981–6990. doi:10.2147/OTT.S214588
12. Ding J, Zhao J, Huan L, et al. LINC00665 increases the malignancy through activating PKR/NF-κB pathway in hepatocellular carcinoma. Hepatology. 2020. doi:10.1002/hep.31195
13. Ji W, Diao YL, Qiu YR, Ge J, Cao XC, Yu Y. LINC00665 promotes breast cancer progression through regulation of the miR-379-5p/LIN28B axis. Cell Death Dis. 2020;11(1):16. doi:10.1038/s41419-019-2213-x
14. Okazaki Y, Furuno M, Kasukawa T, et al.; FANTOM Consortium; RIKEN Genome Exploration. Research Group Phase I & II Team. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573.
15. Zhou JL, Zou L, Zhu T. Long non-coding RNA LINC00665 promotes metastasis of breast cancer cells by triggering EMT. Eur Rev Med Pharmacol Sci. 2020;24(6):3097–3104. doi:10.26355/eurrev_202003_20674
16. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
17. Jiang X, Jiang M, Xu M, Xu J, Li Y. Identification of diagnostic utility and molecular mechanisms of circulating miR-551b-5p in gastric cancer. Pathol Res Pract. 2019;215(5):900–904. doi:10.1016/j.prp.2019.01.035
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.