Back to Journals » OncoTargets and Therapy » Volume 13
LINC00662 Long Non-Coding RNA Knockdown Attenuates the Proliferation, Migration, and Invasion of Osteosarcoma Cells by Regulating the microRNA-15a-5p/Notch2 Axis
Received 7 April 2020
Accepted for publication 10 July 2020
Published 10 August 2020 Volume 2020:13 Pages 7517—7530
DOI https://doi.org/10.2147/OTT.S256464
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Shuheng Liu,1 Xianghai Meng2
1Department of Spine Surgery, Jinan Central Hospital, Cheeloo College of Medicine, Shandong University, Jinan City, Shandong Province, People’s Republic of China; 2Trauma Center, Jinan Central Hospital, Cheeloo College of Medicine, Shandong University, Jinan City, Shandong Province, People’s Republic of China
Correspondence: Xianghai Meng, Jinan City, Shandong Province, People’s Republic of China
Tel + 86- 0531- 55865483
Email [email protected]
Purpose: Osteosarcoma (OS) is a frequently occurring malignancy in children and adolescents. In this study, we aimed to investigate the effects of the long non-coding RNA (lncRNA) LINC00662 (LINC00662) in OS and the underlying molecular mechanism.
Methods: The expression of LINC00662, microRNA-15a-5p (miR-15a-5p), and Notch2 in OS was detected by quantitative real-time polymerase chain reaction (qRT-PCR). The proliferation, migration, and invasion of OS cells were analyzed by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), wound-healing, and transwell assay. The interactions among LINC00662, miR-15a-5p, and Notch2 were determined by dual-luciferase reporter assays. A tumor xenograft model was established in mice for evaluating tumor growth in vivo.
Results: The expression of LINC00662 and Notch2 was found to be upregulated in OS, but the expression of miR-15a-5p was downregulated. The results demonstrated that LINC00662 knockdown attenuated the proliferation, migration, and invasion of OS cells and suppressed tumor growth in mice. The study further demonstrated that LINC00662 directly interacted with miR-15a-5p, and that Notch2 was a target of miR-15a-5p. The inhibition of miR-15a-5p or Notch2 overexpression markedly reversed the suppressive effect of sh-LINC00662 on the proliferation, migration, and invasion of OS cells.
Conclusion: The study demonstrated that LINC00662 could be a potential biomarker for OS therapy, and LINC00662 knockdown suppressed the proliferation, migration, and invasion of OS cells by regulating the miR-15a-5p/Notch2 axis.
Keywords: osteosarcoma, long non-coding RNA LINC00662, microRNA-15a-5p, Notch2, proliferation
Introduction
Osteosarcoma (OS) is a malignant bone tumor1 that frequently occurs in adolescents, and presents with deep pain or other symptoms associated with pathologic fracture.2 Tumor metastasis remains an unfavorable factor for prognosis in patients with low-grade or high-grade OS.3,4 The five-year survival rate of OS continues to be very poor.5 To date, the primary therapeutic agents for OS comprise systemic chemotherapy and surgery for local control.6,7 However, local recurrence following initial treatment is high, and may even require amputation.8 It is therefore essential to explore alternative therapeutic strategies that would be effective in patients with OS.
Long non-coding RNAs (lncRNAs) are essential for the regulation of various cancers, including neuroblastoma,9 breast cancer,10 and OS.11 The lncRNA LINC00662 (LINC00662) is 2085 bp long and is located at 19q11.12 Recent studies have demonstrated that LINC00662 is involved in numerous cellular and biological processes of cancer cells. For instance, LINC00662 enhances the growth of gastric cancer cells by regulating the Hippo-YAP1 signaling pathway,13 and promotes the growth of oral squamous carcinoma cells.14 LINC00662 accelerates tumorigenesis and the progression of colorectal cancer by modulating the miR-497-5p/AVL9 axis.15 However, despite numerous studies on LINC00662, the precise role of LINC00662 in the progression of OS remains unclear.
Certain microRNAs (miRNAs) have been shown to have anti-cancer effects in OS. For instance, miRNA-218 decreases the migration and invasion of OS cells by inhibiting the expression of T-lymphoma and metastasis 1 (TIAM1).16 MiRNA-646 inhibits the metastasis of OS cells by downregulating fibroblast growth factor 2 (FGF2).17 MiRNA-15a suppresses the tumorigenesis of OS cells by targeting tumor necrosis factor, alpha-induced protein 1 (TNFAIP1).18 Studies by Jiang et al (2017) and Wang et al (2018) demonstrated that the miRNAs that interact with lncRNAs are related to the oncogenic processes in OS.19,20 However, the precise regulatory relationship between LINC00662 and miR-15a-5p in OS remains to be elucidated.
Notch2 is an important mammalian Notch receptor that participates in the cellular processes of various cancers, including OS.21,22 A previous study demonstrated that Notch2 is overexpressed in tissue specimens obtained from patients with OS23 and can serve as a therapeutic target for OS.24 Additionally, Zhou et al (2018) revealed that the SNHG12 lncRNA promotes tumorigenesis in OS by sponging miR-195-5p, which subsequently upregulates Notch2 expression.25 Nevertheless, the potential regulatory mechanism of LINC00662 in OS, mediated via the miR-15a-5p/Notch2 axis, remains to be elucidated.
In this study, we investigated the effects of LINC00662 on OS and evaluated the relationship between LINC00662 and miR-15a-5p. The results demonstrated that the expression of LINC00662 increased in OS, and LINC00662 acted as a competing endogenous RNA (ceRNA) for the tumor suppressor miRNA, miR-15a-5p, which upregulated Notch2 expression. In this study, we identified the role of LINC00662 in the tumorigenesis of OS. Additionally, our results confirmed the interactions among LINC00662, miR-15a-5p, and Notch2, thus providing novel information for the development of alternative therapeutic strategies for OS. The results of our study may aid in the identification of effective therapeutic targets for OS.
Materials and Methods
Tissue Samples
A total of 57 patients with OS, of which 28 were males and 29 were females, and aged between 15.36 ± 1.31 years, were selected from our hospital between January 2017 and October 2018. The patients had not received previous treatment with radiotherapy or chemotherapy. Samples of OS tissues and adjacent noncancerous tissues were collected from the patients. Based on the median expression of LINC00662, our patient cohort was categorized into high LINC00662 (n=29) and low LINC00662 groups (n=28). The study was approved by the ethics committee of Jinan Central Hospital, Cheeloo College of Medicine, Shandong University. Written informed consent was obtained from all the patients and parental consent was obtained in cases where the patient was less than 18 years old. The study was conducted in accordance with the Declaration of Helsinki.
Cell Culture
Four human OS cell lines, HOS, MG63, SJSA-1, and U2OS, and the human fetal osteoblastic cell line, hFOB, were obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen) at 37°C.
Cellular Transfection
The miR-negative control (NC), miR-15a-5p mimics, miR-15a-5p inhibitor, pcDNA3.1 Notch2 (pcDNA-Notch2), and pcDNA-NC were synthesized by Invitrogen. The lentiviral vector, short hairpin-LINC00662, and sh-NC were synthesized by GenePharma (Shanghai, China). The MG63 and U2OS cells were cultured to a confluence of 80%, and were subsequently transfected or co-transfected with the aforementioned agents using Lipofectamine 3000 (Invitrogen).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) and Western Blotting
We subsequently performed qRT-PCR and Western blotting as previously described.26 The primers are enlisted in Table 1. GAPDH, U6, and β-actin were used to normalize the expression levels of LINC00662, miR-15a-5p, and Notch2, respectively. The antibodies used for Western blot analysis included the anti-Notch2 primary antibody (1:1000, SAB4502021MSDS, St. Louis, MO, USA), anti-β-actin antibody (1:5000, SAB2701711MSDS, Sigma), and HRP-conjugate secondary antibody (1:5000, 12–348MSDS, Sigma). The protein bands were visualized by exposure to enhanced chemiluminescence solution, and quantified by ImageLab software (Bio-Rad, Hercules, CA, USA).
 |
Table 1 Primers Sequences |
Tumor Formation in Mice
Male BALB/c nude mice (4-week-old) were purchased from Beijing HFK Bioscience Co., Ltd. (China, Beijing). The mice were housed at 22–25°C in an atmosphere of 50–60% relative humidity, and fed on a normal diet. U2OS cells (7×106 cells in 200 μL of sterile Dulbecco’s phosphate-buffered saline) were transfected with sh-NC and sh-LINC00662, and injected into the abdominal cavity of the mice in the sh-NC and sh-LINC00662 groups, respectively (n=4). The tumor volumes were measured on a weekly basis, commencing from one week post-inoculation. The tumor volumes were calculated according to the following formula: (A.B2)/2 (where A is the longest diameter in millimeters, and B is the shortest diameter in millimeters). After the last measurement in the 4th week, the mice were anesthetized with 50 mg/kg pentobarbital sodium, and the intact tumors were exfoliated and weighed. The animal experiments were approved by the ethics committee of Jinan Central Hospital, Cheeloo College of Medicine, Shandong University, and were performed in accordance with the institutional guide for the care and use of laboratory animals (National Institutes of Health, USA).
3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
MG63 and U2OS cells were seeded into 96-well plates at a density of 2×103 cells/well. Following the addition of 20 μL MTT (Sigma) at specified time points of 0, 24, 48, 72, and 96 h, the cells were incubated for 4 h at 37°C in an atmosphere of 5% CO2. Subsequently, 150 μL of dimethyl sulfoxide (DMSO) was added for dissolving the formazan after discarding the supernatant. The absorbance was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Wound Healing Assay
MG63 and U2OS cells were seeded into a 6-well plate and cultured until they reached a confluence of 80% in DMEM supplemented with 10% FBS. A sterile pipette tip was used to draw a vertical line on the surface of the cell culture plate. The cells were then cultured in DMEM without FBS. The width of the scratch was measured at 0 and 24 h. The cells were observed under a light microscope and the wound healing rate was subsequently calculated.
Transwell Assay
The transwell assay was used to determine cell invasion using transwell chambers of 8 nm pore size (Corning Inc., Corning, NY, USA). MG63 and U2OS cells (2 × 105), cultured in serum-free medium, were added to the upper transwell chambers pre-coated with matrigel (Sigma). RPMI-1640 (Invitrogen) medium, containing serum, was added to the lower chambers. The cells were subsequently removed from the upper chambers with a cotton swab after 48 h of incubation at 37°C, while the cells in the lower chambers were fixed in methanol and stained with 0.5% crystal violet for 15 min. Cells from five random fields were photographed and counted for analysis.
RNA Immunoprecipitation (RIP) Assay
The RIP assay was performed using the EZ-Magna RIP kit (Millipore, Billerica, MA, USA). MG63 and U2OS cells, at a confluence of 80%, were incubated with RIP buffer containing magnetic beads conjugated with human anti-Ago2 antibody (Millipore). IgG (Millipore) was used as the control. The immunoprecipitated RNAs were subsequently isolated by Proteinase K. The purified RNAs were finally analyzed by qRT-PCR.
Bioinformatics-Based Prediction and Analyses
We used two independent online tools, LncBase v2.0 and starBase v3.0, for prediction and identification of the miRNAs that target LINC00662. The miR-15a-5p miRNA was simultaneously identified by both the prediction tools. MiR-15a-5p is a well-known anti-oncogene, which downregulated in various cancers. The miR-15a-5p miRNA was therefore selected as the candidate miRNA for subsequent studies. We additionally employed the online tools, targetScan, miRDB, and miRTarBase, for predicting the possible target genes of miR-15a-5p. The Notch2 gene was simultaneously identified by all the three prediction tools. Notch2 is a well-known oncogene and is a therapeutic target for OS. We therefore selected Notch2 for subsequent studies.
Dual-Luciferase Reporter Assay
LINC00662-Wt, LINC00662-Mut, Notch2-Wt, and Notch2-Mut were cloned and combined with psiCHECK-2 vectors (Promega, Madison, WI, USA). They were subsequently co-transfected with miR-NC and miR-15a-5p mimics into MG63 and U2OS cells by incubating with Lipofectamine 3000 for 48 h, and detected by the dual-luciferase reporter assay system (Promega).
Statistical Analyses
All the statistical analyses were performed on GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). The data are presented as the mean ± standard deviation (SD). The differences between two groups or among multiple groups were assessed by Student’s t-test or one-way ANOVA followed by Tukey’s post hoc test. The significance of the correlations was determined by Pearson’s correlation analysis. P values < 0.05 were considered to be statistically significant.
Results
LINC00662 Expression Increased in OS
The expression of LINC00662 was markedly increased in the tumor tissues in comparison to that of the adjacent noncancerous tissues obtained from the patients with OS (P < 0.001, Figure 1A). Compared to tumor-node-metastasis (TNM) I/II, the expression of LINC00662 was markedly increased in TNM III/IV (P < 0.001, Figure 1B). Besides, the expression of LINC00662 in the metastatic tumors was visibly increased in comparison to that of the non-metastatic tumors (P < 0.001, Figure 1C). Gene Expression Profiling Interactive Analysis (GEPIA) with The Cancer Genome Atlas (TCGA) revealed that the overall survival of patients with OS with high LINC00662 expression was markedly shorter in comparison to that of patients with low LINC00662 expression (P < 0.05, Figure 1D). The clinicopathological features revealed that the degree of metastasis and World Health Organization (WHO) Grade of patients with the low LINC00662 expression were significantly different from those of patients with high LINC00662 expression (P < 0.05, Table 2).
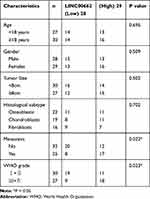 |
Table 2 Correlation Between LINC00662 Expression and Clinicopathological Features in Osteosarcoma (OS) Patients |
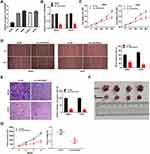
LINC00662 Knockdown Inhibited Tumorigenesis of OS Cells
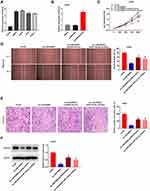
The expression of LINC00662 visibly increased in HOS, MG63, SJSA-1, and U2OS cells, in comparison to that in hFOB cells (P < 0.01, Figure 2A). Owing to the relative high expression of LINC00662, the MG63 and U2OS cells were selected for further experimentation. LINC00662 knockdown was first performed by transfecting MG63 and U2OS cells with sh-LINC00662. Subsequently, qRT-PCR was performed, which revealed that the expression of LINC00662 was downregulated in cells transfected with sh-LINC00662 (P < 0.01, Figure 2B). The MTT assay revealed that sh-LINC00662 significantly reduced the proliferation of MG63 and U2OS cells after 72 and 96 h of culture (P < 0.05, Figure 2C). Additionally, wound healing and the invasion rate of MG63 and U2OS cells were markedly reduced in the sh-LINC00662 group in comparison to those of the cells in the sh-NC group (P < 0.01, Figure 2D and E). In order to elucidate the function of LINC00662 in vivo, U2OS cells transfected with sh-NC or sh-LINC00662 were intraperitoneally injected into mice. As expected, LINC00662 knockdown reduced tumor volume and tumor weight (P < 0.01, Figure 2F and G).
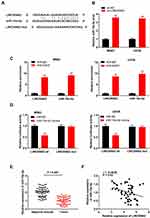
LINC00662 Expression Was Negatively Correlated with miR-15a-5p Expression
The binding site of miR-15a-5p, as predicted by starBase v3.0, was at the 3′-UTR of LINC00662 (Figure 3A). As depicted in Figure 3B, the downregulation of LINC00662 significantly increased the expression of miR-15a-5p (P < 0.01). The results of RIP assay indicated that the relative enrichment of LINC00662 and miR-15a-5p in MG63 and U2OS cells were strikingly abundant in the anti-Ago2 group in comparison to that of the anti-IgG group (P < 0.01, Figure 3C). The relative luciferase activity was markedly decreased in MG63 and U2OS cells co-transfected with miR-15a-5p mimics and LINC00662 Wt, compared to that of cells co-transfected with miR-NC and LINC00662 Wt (P < 0.01, Figure 3D). The expression of miR-15a-5p in the tumor tissues was markedly reduced in comparison to that in the adjacent noncancerous tissues obtained from patients with OS (P < 0.001, Figure 3E). There was a negative correlation between the expression of LINC00662 and miR-15a-5p in the OS tissues (N = 57, r = −0.5569, P < 0.001, Figure 3F).
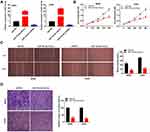
MiR-15a-5p Inhibited the Proliferation, Migration, and Invasion of OS Cells
MiR-15a-5p was overexpressed by transfecting with miR-15a-5p mimics, and inhibited by transfecting with the miR-15a-5p inhibitor (P < 0.01, Figure 4A). Transfection with miR-15a-5p mimics markedly reduced the proliferation of MG63 and U2OS cells after 72 and 96 h of culture (P < 0.01, Figure 4B). Additionally, miR-15a-5p significantly reduced wound healing and the invasion rate of MG63 and U2OS cells (P < 0.01, Figure 4C and D).
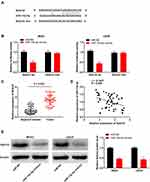
Notch2 Was Directly Targeted by miR-15a-5p
The binding site for miR-15a-5p, as predicted by targetScan, was at the 3′-UTR of Notch2 (Figure 5A). The relative luciferase activity was visibly decreased in MG63 and U2OS cells co-transfected with miR-15a-5p mimics and Notch2 Wt, in comparison to that of cells co-transfected with miR-NC and Notch2 Wt (P < 0.01, Figure 5B). Furthermore, the expression of Notch2 in the tumor tissues was markedly increased in comparison to that of the adjacent normal tissues obtained from the patients with OS (P < 0.001, Figure 5C). There was a negative correlation between the expression of Notch2 and miR-15a-5p in the OS tissues (N = 57, r = −0.5886, P < 0.001, Figure 5D). The results of Western blotting revealed that transfection with miR-15a-5p mimics downregulated the expression of the Notch2 protein in MG63 and U2OS cells (P < 0.01, Figure 5E).
LINC00662 Knockdown Attenuated the Proliferation, Migration, and Invasion of OS Cells by Targeting miR-15a-5p/Notch2 Axis
The expression of Notch2 visibly increased in MG63, U2OS, HOS, and SJSA-1 cells, in comparison to that of hFOB cells (P < 0.01, Figure 6A). Owing to the relative high expression of Notch2, the U2OS cells were selected for further experimentation. The expression of Notch2 was upregulated following transfection with pcDNA-Notch2 (P < 0.01, Figure 6B). The proliferation, wound healing rate, and rate of invasion of U2OS cells were markedly reduced in the sh-LINC00662 group in comparison to those of the sh-NC group (P < 0.01). The overexpression of Notch2 or the inhibition of miR-15a-5p markedly reversed the suppressive effect of sh-LINC00662 on the proliferation, wound healing rate, and rate of invasion of U2OS cells (P < 0.05, Figure 6C–E). Furthermore, sh-LINC00662 markedly downregulated the expression of Notch2 protein in U2OS cells (P < 0.01). Rescue assays revealed that Notch2 overexpression or miR-15a-5p inhibition markedly rescued the inhibitory effect of sh-LINC00662 on the expression of the Notch2 protein in U2OS cells (P < 0.05, Figure 6F).
Discussion
The overexpression of lncRNAs is associated with the tumorigenesis of OS.27,28 The expression of ncRNAs such as lncRNA TUG1,29 lncRNA BANCR,30 and lncRNA ODRUL31 increases in patients with OS. The results of this study demonstrated that the expression of LINC00662 was higher in the tumor tissues than that in the adjacent noncancerous tissues obtained from the patients with OS. Additionally, the expression of LINC00662 was found to be correlated with degree of metastasis and WHO grade in patients with OS, and a high expression of LINC00662 conferred a survival disadvantage to the patients with OS. The function of LINC00662 was found to be similar to that of the lncRNAs reported in previous studies. The high expression of the CBR3-AS1 lncRNA is associated with distant metastasis in patients with OS.32 The high expression of the UCA1 lncRNA is markedly related to high tumor grade in patients with OS.33 The upregulation of the PVT1 lncRNA decreases the survival rate of patients with OS.34 In summary, the results of this study suggested that the expression of LINC00662 is upregulated in patients with OS, and could be associated with the development of OS.
Previous studies have confirmed that LINC00662 partakes in the cellular proliferation, invasion, and migration of various cancers. LINC00662 enhances the invasion of lung cancer cells by interacting with LIN28.35 The silencing of LINC00662 alleviates the proliferation, invasion, and migration of prostate cancer cells by regulating miR-34a.36 The inhibition of LINC00662 attenuates the growth of acute myeloid leukemia cells by sponging miR-340-5p.37 Notably, LINC00662 knockdown suppresses the growth, migration, and invasion of colorectal cancer cells via regulating the miR-497-5p/AVL9 axis.15 In the present study, LINC00662 knockdown reduced the proliferation, migration, and invasion of OS cells. The function of LINC00662 identified herein was similar to the results of the aforementioned studies, and indicates that LINC00662 could serve as a promising therapeutic target for OS. Wang et al (2020) demonstrated that the inhibition of LINC00662 attenuates the growth of chordomas in vivo.38 The study by Tian et al (2020) revealed that LINC00662 silencing markedly inhibits the growth of hepatic tumors in mice.39 In this study, sh-LINC00662 markedly attenuated tumor growth in mice. The study further illustrates that LINC00662 knockdown attenuates the tumorigenesis of OS in vivo.
It has been documented that non-coding RNAs, including lncRNAs and miRNAs, are involved in the progression of OS. For instance, the B4GALT1-AS1 lncRNA recruits HuR to increase the stemness and migration of OS cells by elevating the transcriptional activity of YAP.40 The RNA-binding protein, Pumilio2 (PUM2), attenuates the progression of OS by partially and competitively binding to the 3′-UTR of StAR-related lipid transfer domain 13 (STARD13), together with miR-50-3p and miR-9.41 Interestingly, lncRNAs can serve as ceRNAs or miRNA sponges for regulating the expression of target mRNAs in OS.42–44 The SNHG15 lncRNA facilitates the proliferation of OS cells by sponging miR-141.45 The LINC00963 lncRNA contributes to the growth of OS cells by inhibiting the miR-204-3p/FN1 axis.46 The C2dat1 lncRNA accelerates tumorigenesis and metastasis by suppressing the expression of miR-34a-5p in OS.47 In this study, we confirmed that miR-15a-5p is a target of LINC00662. MiR-15a-5p is a tumor suppressor, and its expression is downregulated in endometrial cancer,48 melanoma,49 and papillary thyroid cancer.50 The results of this study demonstrated that the expression of miR-15a-5p is downregulated in the tumor tissues and cells of OS, and its expression is negatively correlated to that of LINC00662. Studies by Cai et al (2012), Tian et al (2015), and Leng et al (2018) have demonstrated that miR-15a induces cell cycle arrest and promotes apoptosis in OS by downregulating the expression of CCND1,51 and attenuates the growth of OS cells by targeting TNFAIP118 or Bcl-2.52 In this study, we observed that miR-15a-5p inhibited the proliferation, migration, and invasion of OS cells, and the inhibition of miR-15a-5p reversed the anti-tumor effect of LINC00662 knockdown in OS cells. These results indicated that LINC00662 knockdown can inhibit tumorigenesis by upregulating miR-15a-5p in OS cells.
The expression of Notch2 is generally upregulated in various tumors, including embryonal brain tumor,53 pancreatic cancer,54 and OS.55 Similar to the results of previous studies, we observed that the expression of Notch2 was markedly increased in the tumor tissues of patients with OS. Notch2 is involved in the progression of various cancers. For instance, Notch2 accelerates the formation of cholangiocarcinoma.56 The upregulation of Notch2 expression contributes to the development of breast cancer.57 Therefore, Notch2 could be involved in the progression of OS progression, as it is a target gene for miRNAs. MiR-1296-5p reduces tumorigenesis in OS by targeting Notch2.58 Here, we identified Notch2 as a target of the miR-15a-5p miRNA, and the expression of Notch2 was found to be negatively correlated to that of miR-15a-5p. The feedback data revealed that the overexpression of Notch2 reversed the anti-tumor effect of sh-LINC00662 in OS cells, suggesting that sh-LINC00662 may exert its anti-tumor effect by regulating the miR-15a-5p/Notch2 axis in OS.
In conclusion, the expression of LINC00662 was found to be upregulated in OS tissues and cells. We identified that miR-15a-5p is a target of LINC00662, and that Notch2 is a target of miR-15a-5p. The study further demonstrated that LINC00662 knockdown attenuates the proliferation, migration, and invasion of OS cells by regulating the miR-15a-5p/Notch2 axis, and inhibits tumor growth in mice. These results indicate that LINC00662 could be a promising therapeutic target for OS.
Data Sharing Statement
All data are available through corresponding author, Xianghai Meng.
Ethics Approval and Consent to Participate
The study was approved by the ethics committee of Jinan Central Hospital, Cheeloo College of Medicine, Shandong University. Written informed consent was obtained from all the patients and parental consent was obtained in cases where the patient was less than 18 years old. The study was conducted in accordance with the Declaration of Helsinki.
The animal experiments were approved by the ethics committee of Jinan Central Hospital, Cheeloo College of Medicine, Shandong University, and were performed in accordance with the institutional guide for the care and use of laboratory animals (National Institutes of Health, USA).
Acknowledgments
Lin Chen, Xi Wang, and Qingguo Zhang made contributions in the revising of the manuscript. Fei Gao helped prepare the manuscript.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The Authors declare that they have no conflicts of interest to disclose.
References
1. Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma overview. Rheumatol Ther. 2017;4(1):25–43. doi:10.1007/s40744-016-0050-2
2. Franceschini N, Cleton-Jansen A-M, Bovée JV. Bone: conventional osteosarcoma. Atlas Genet Cytogenet Oncol Haematol. 2019;(8). doi:10.4267/2042/70484.
3. Berner K, Johannesen TB, Bruland ØS. Clinical epidemiology of low-grade and dedifferentiated osteosarcoma in Norway during 1975 and 2009. Sarcoma. 2015;2015:1–9. doi:10.1155/2015/917679
4. Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high-grade osteosarcoma using the surveillance, epidemiology, and end results (SEER) program database. Cancer Epidemiol. 2015;39(4):593–599. doi:10.1016/j.canep.2015.05.001
5. Wahidah T, Khattak MN, Wan-Arfah N, Naing NN. Five-year survival of osteosarcoma patients in Hospital Universiti Sains Malaysia (HUSM): an eleven year review. Res J Pharm Technol. 2018;11(8):3534–3542. doi:10.5958/0974-360X.2018.00653.4
6. Ferrari S, Serra M. An update on chemotherapy for osteosarcoma. Expert Opin Pharmacother. 2015;16(18):2727–2736. doi:10.1517/14656566.2015.1102226
7. Durfee RA, Mohammed M, Luu HH. Review of osteosarcoma and current management. Rheumatol Ther. 2016;3(2):221–243. doi:10.1007/s40744-016-0046-y
8. Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18(1):39–50. doi:10.1080/14737140.2018.1413939
9. Zhang J, Li WY, Yang Y, et al. LncRNA XIST facilitates cell growth, migration and invasion via modulating H3 histone methylation of DKK1 in neuroblastoma. Cell Cycle. 2019;18(16):1882–1892. doi:10.1080/15384101.2019.1632134
10. Shi S-J, Wang L-J, Yu B, Li Y-H, Jin Y, Bai X-Z. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6(13):11652. doi:10.18632/oncotarget.3457
11. Sun X-H, Yang L-B, Geng X-L, Wang R, Zhang Z-C. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8(3):2994.
12. Team MGCP. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99(26):16899–16903. doi:10.1073/pnas.242603899
13. Liu Z, Yao Y, Huang S, et al. LINC00662 promotes gastric cancer cell growth by modulating the hippo-YAP1 pathway. Biochem Biophys Res Commun. 2018;505(3):843–849. doi:10.1016/j.bbrc.2018.09.191
14. Xu D, Chen Y, Yuan C, Zhang S, Peng W. Long non-coding RNA LINC00662 promotes proliferation and migration in oral squamous cell carcinoma. Onco Targets Ther. 2019;12:647. doi:10.2147/OTT.S188691
15. Wang H, Yu M, Hu W, et al. Linc00662 promotes tumorigenesis and progression by regulating miR-497-5p/AVL9 axis in colorectal cancer. Front Genet. 2020;10:1385. doi:10.3389/fgene.2019.01385
16. Jin J, Cai L, Liu Z-M, Zhou X-S. miRNA-218 inhibits osteosarcoma cell migration and invasion by down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev. 2013;14(6):3681–3684. doi:10.7314/APJCP.2013.14.6.3681
17. Sun X-H, Geng X-L, Zhang J, Zhang C. miRNA-646 suppresses osteosarcoma cell metastasis by downregulating fibroblast growth factor 2 (FGF2). Tumor Biol. 2015;36(3):2127–2134. doi:10.1007/s13277-014-2822-z
18. Tian X, Zhang J, Yan L, Dong J-M, Guo Q. MiRNA-15a inhibits proliferation, migration and invasion by targeting TNFAIP1 in human osteosarcoma cells. Int J Clin Exp Pathol. 2015;8(6):6442.
19. Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi:10.1016/j.canlet.2017.06.009
20. Wang Y, Kong D. Knockdown of lncRNA MEG3 inhibits viability, migration, and invasion and promotes apoptosis by sponging miR‐127 in osteosarcoma cell. J Cell Biochem. 2018;119(1):669–679. doi:10.1002/jcb.26230
21. Wang C, Li Q, Liu F, et al. Notch2 as a promising prognostic biomarker for oesophageal squamous cell carcinoma. Sci Rep. 2016;6(1):25722. doi:10.1038/srep25722
22. Li R, Zhang W, Cui J, et al. Targeting BMP9-promoted human osteosarcoma growth by inactivation of notch signaling. Curr Cancer Drug Targets. 2014;14(3):274–285. doi:10.2174/1568009614666140305105805
23. Tanaka M, Setoguchi T, Hirotsu M, et al. Inhibition of notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer. 2009;100(12):1957–1965.
24. Kannan S, Livingston JA, Roth M, et al. Notch2 Inhibition as a Therapeutic Intervention in Osteosarcoma. AACR; 2019.
25. Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. 2018;495(2):1822–1832. doi:10.1016/j.bbrc.2017.12.047
26. Yan L, Wu X, Yin X, Du F, Ding X, Ding X. LncRNA CCAT2 promoted osteosarcoma cell proliferation and invasion. J Cell Mol Med. 2018;22(5):2592–2599. doi:10.1111/jcmm.13518
27. Chen X, Zhou Y, Liu S, et al. LncRNA TP73‐AS1 predicts poor prognosis and functions as oncogenic lncRNA in osteosarcoma. J Cell Biochem. 2019;120(2):2569–2575. doi:10.1002/jcb.27556
28. Zhao H, Hou W, Tao J, et al. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway. Am J Transl Res. 2016;8(8):3503.
29. Yun-Bo F, Xiao-Po L, Xiao-Li L, Guo-Long C, Pei Z, Fa-Ming T. LncRNA TUG1 is upregulated and promotes cell proliferation in osteosarcoma. Open Med. 2016;11(1):163–167. doi:10.1515/med-2016-0031
30. Peng Z, Lu R, Xiao D, Xiao Z. Increased expression of the lncRNA BANCR and its prognostic significance in human osteosarcoma. Genet Mol Res. 2016;15(1). doi:10.4238/gmr.15017480
31. Zhu K-P, Ma X-L, Zhang C-L. LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 Axis. Mol Ther. 2017;25(10):2383–2393. doi:10.1016/j.ymthe.2017.06.027
32. Zhang Y, Meng W, Cui H. LncRNA CBR3-AS1 predicts unfavorable prognosis and promotes tumorigenesis in osteosarcoma. Biomed Pharmacother. 2018;102:169–174. doi:10.1016/j.biopha.2018.02.081
33. Li W, Xie P, Ruan W-H. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol. 2016;5(2):80–85. doi:10.1016/j.jbo.2016.05.003
34. Zhou Q, Chen F, Zhao J, et al. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget. 2016;7(50):82620. doi:10.18632/oncotarget.13012
35. Gong W, Su Y, Liu Y, Sun P, Wang X. Long non-coding RNA Linc00662 promotes cell invasion and contributes to cancer stem cell-like phenotypes in lung cancer cells. J Biochem. 2018;164(6):461–469. doi:10.1093/jb/mvy078
36. Li N, Zhang L, Qiao Y, Song R. Long noncoding RNA LINC00662 functions as miRNA sponge to promote the prostate cancer tumorigenesis through targeting miR-34a. Eur Rev Med Pharmacol Sci. 2019;23(9):3688–3698. doi:10.26355/eurrev_201905_17792
37. Liu Y, Gao X, Tian X. High expression of long intergenic non-coding RNA LINC00662 contributes to malignant growth of acute myeloid leukemia cells by upregulating ROCK1 via sponging microRNA-340-5p. Eur J Pharmacol. 2019;859:172535. doi:10.1016/j.ejphar.2019.172535
38. Wang CB, Wang Y, Wang JJ, Guo XL. LINC00662 triggers malignant progression of chordoma by the activation of RNF144B via targeting miR-16-5p. Eur Rev Med Pharmacol Sci. 2020;24(3):1007–1022. doi:10.26355/eurrev_202002_20151
39. Tian X, Wu Y, Yang Y, et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/beta-catenin signaling. Mol Oncol. 2020;14(2):462–483. doi:10.1002/1878-0261.12606
40. Li Z, Wang Y, Hu R, Xu R, Xu W. LncRNA B4GALT1-AS1 recruits HuR to promote osteosarcoma cells stemness and migration via enhancing YAP transcriptional activity. Cell Prolif. 2018;51(6):e12504. doi:10.1111/cpr.12504
41. Hu R, Zhu X, Chen C, Xu R, Li Y, Xu W. RNA-binding protein PUM2 suppresses osteosarcoma progression via partly and competitively binding to STARD13 3ʹUTR with miRNAs. Cell Prolif. 2018;51(6):e12508. doi:10.1111/cpr.12508
42. Liao S, Xing S, Ma Y. LncRNA SNHG16 sponges miR-98-5p to regulate cellular processes in osteosarcoma. Cancer Chemother Pharmacol. 2019;83(6):1065–1074. doi:10.1007/s00280-019-03822-5
43. Luo W, He H, Xiao W, et al. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget. 2016;7(34):54733. doi:10.18632/oncotarget.10752
44. Kong D, Wang Y. Knockdown of lncRNA HULC inhibits proliferation, migration, invasion, and promotes apoptosis by sponging miR‐122 in osteosarcoma. J Cell Biochem. 2018;119(1):1050–1061. doi:10.1002/jcb.26273
45. Liu K, Hou Y, Liu Y, Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J Biomed Sci. 2017;24(1):46. doi:10.1186/s12929-017-0353-9
46. Zhou Y, Yin L, Li H, Liu L-H, Xiao T. The LncRNA LINC00963 facilitates osteosarcoma proliferation and invasion by suppressing miR-204-3p/FN1 axis. Cancer Biol Ther. 2019;20(8):1141–1148. doi:10.1080/15384047.2019.1598766
47. Jia D, Niu Y, Li D, Liu Z. lncRNA C2dat1 promotes cell proliferation, migration, and invasion by targeting miR-34a-5p in osteosarcoma cells. Oncol Res. 2018;26(5):753–764. doi:10.3727/096504017X15024946480113
48. Wang Z, Wan X, Sang G, Zhao J, Zhu Q, Wang D. miR-15a-5p suppresses endometrial cancer cell growth via Wnt/β-catenin signaling pathway by inhibiting WNT3A. Eur Rev Med Pharmacol Sci. 2017;21(21):4810–4818.
49. Alderman C, Yang Y. The anti-melanoma activity and oncogenic targets of hsa-miR-15a-5p. RNA Dis. 2016;3(4).
50. Jiang L, Wu Z, Meng X, Chu X, Huang H, Xu C. LncRNA HOXA-AS2 facilitates tumorigenesis and progression of papillary thyroid cancer by modulating the miR-15a-5p/HOXA3 axis. Hum Gene Ther. 2019;30(5):618–631. doi:10.1089/hum.2018.109
51. Cai C-K, Zhao G-Y, Tian L-Y, et al. miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol Rep. 2012;28(5):1764–1770. doi:10.3892/or.2012.1995
52. Leng J, Song Q, Zhao Y, Wang Z. miR‑15a represses cancer cell migration and invasion under conditions of hypoxia by targeting and downregulating Bcl‑2 expression in human osteosarcoma cells. Int J Oncol. 2018;52(4):1095–1104. doi:10.3892/ijo.2018.4285
53. Fan X, Mikolaenko I, Elhassan I, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64(21):7787–7793. doi:10.1158/0008-5472.CAN-04-1446
54. Mazur PK, Einwächter H, Lee M, et al. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2010;107(30):13438–13443. doi:10.1073/pnas.1002423107
55. Hughes DP. How the NOTCH pathway contributes to the ability of osteosarcoma cells to metastasize. In: Pediatric and Adolescent Osteosarcoma. Springer; 2009:479–496.
56. Dill MT, Tornillo L, Fritzius T, et al. Constitutive Notch2 signaling induces hepatic tumors in mice. Hepatology. 2013;57(4):1607–1619. doi:10.1002/hep.26165
57. Fu Y-P, Edvardsen H, Kaushiva A, et al. NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. Mol Cancer. 2010;9(1):113. doi:10.1186/1476-4598-9-113
58. Wang L, Hu K, Chao Y, Wang X. MicroRNA‐1296‐5p suppresses the proliferation, migration, and invasion of human osteosarcoma cells by targeting NOTCH2. J Cell Biochem. 2020;121(2):2038–2046. doi:10.1002/jcb.29438
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.