Back to Journals » OncoTargets and Therapy » Volume 12
LINC00565 promotes proliferation and inhibits apoptosis of gastric cancer by targeting miR-665/AKT3 axis
Authors Hu J, Ni G, Mao L, Xue X, Zhang J, Wu W, Zhang S, Zhao H, Ding L, Wang L
Received 2 October 2018
Accepted for publication 28 May 2019
Published 24 September 2019 Volume 2019:12 Pages 7865—7875
DOI https://doi.org/10.2147/OTT.S189471
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Jianghong Hu,1,* Guohua Ni,2,* Ling Mao,2 Xianglong Xue,1 Jijie Zhang,2 Weixia Wu,2 Shaoru Zhang,3 Hong Zhao,3 Lifang Ding,2 Lihui Wang3
1Department of Gastroenterology, Danyang People’s Hospital of Jiangsu Province and Danyang Hospital Affiliated to Nantong University, Danyang, Jiangsu 212300, People’s Republic of China; 2Department of Oncology, Danyang People’s Hospital of Jiangsu Province and Danyang Hospital Affiliated to Nantong University, Danyang, Jiangsu 212300, People’s Republic of China; 3Central Laboratory, Danyang People’s Hospital of Jiangsu Province and Danyang Hospital Affiliated to Nantong University, Danyang, Jiangsu 212300, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hong Zhao; Lihui Wang
Danyang People’s Hospital of Jiangsu Province, 2 Xinmin West Road, Danyang 212300, People’s Republic of China
Tel +86 511 8658 0096
Fax +86 511 8655 3198
Email [email protected]; [email protected]
Background: Numerous studies have shown that long noncoding RNA (lncRNA) is involved in gastric cancer (GC). A relevant microarray containing gastric cancer-related lncRNAs was downloaded from The Cancer Genome Atlas database.
Methods: qRT-PCR was used to analyze LINC00565 and AKT3 expression in tumor tissues and cell lines. Proliferative, colony formation and apoptotic abilities of GC cells after transfection of sh-LINC00565 were determined by CCK-8, colony formation assay and flow cytometry, respectively. RIP was enrolled to detect the interaction between LINC00565, AKT3 and miR-665. Dual luciferase assay was used to confirm the relation between miR-665 and LINC00565 and AKT3.
Results: Expression level of LINC00565 in GC tissue was highly expressed in GC, which was negatively correlated to prognosis of GC patients. The results showed that knockdown of LINC00565 decreased proliferative and colony formation abilities, and induced apoptosis of GC cells. Pearson analysis showed that LINC00565 was positively correlated with AKT3. Besides, AKT3 was significantly up-regulated in GC. In addition, knockdown of LINC00565 down-regulated AKT3. In order to explore the mechanism, we found that miR-665 could bind to LINC00565 by bioinformatics. Dual-luciferase reporter gene assay and RIP assay both verified the binding relationship between miR-665 and AKT3. Finally, rescue experiments were carried out to explore whether AKT3 could reverse the anti-cancer effect of low-level LINC00565 on GC development.
Conclusion: In summary, the expression of LINC00565 is upregulated in GC. LINC00565 can be used as the sponge of miR-665 to up-regulate the expression of AKT3, thus promoting the progression of GC.
Keywords: CeRNA, GC, LINC00565, MicroRNA-665, AKT3, proliferation, apoptosis
Introduction
The incidence of gastric cancer (GC) ranks the fifth, and its mortality rate ranks the third globally.1 Clinical treatments of GC have been continuously improved, including the development of new chemotherapy drugs and application of tumor immunotherapy.2–4 However, the 5-year survival of GC is still unsatisfactory.5 GC is a type of tumor disease with high heterogeneity. Most GC patients have already progressed into the advanced stage of GC due to difficulties in the early-stage diagnosis.6 Clarify the molecular mechanism underlying the occurrence and progression of GC has important clinical value for improving its detection, diagnosis and treatment efficacies.
As the progression in sequencing technology, analyses of genomes and transcriptomes have entered a new era. Current studies have shown that 85% of the entire human genome can be transcribed.7,8 However, RNA transcripts with protein-encoding function are much smaller than the number of transcribed RNAs, indicating that a large number of transcripts do not encode proteins, which are called non-coding RNAs (ncRNAs). MicroRNAs are ncRNA with 21–24 nucleotides in length.9–11 The biological characteristics and functions of microRNAs have been extensively studied, and their gene regulation has been deeply elucidated. However, biological characteristics and functions of lncRNAs are less studied.
It is believed that lncRNAs are the basic elements of transcriptional regulation triggering explorations on the molecular mechanism and functional roles of lncRNAs.12,13 Existing studies have shown that some lncRNAs are associated with normal body development and diseases. LncRNAs are involved in a variety of biological processes, including embryonic stem cells, differentiation of helper T cells, autophagy, myocardial infarction, cellular senescence, apoptosis and metastasis of cancer cells, and chemotherapy resistance.14,15 LncRNAs have been found to be dysregulated in various cancer diseases, which are related to cancer recurrence and poor prognosis.16,17 Up to now, functional characteristics of several lncRNAs in GC have been well elucidated, such as HOTAIR, ANRIL, H19, GAPLINC, PVT1, and LINC00152.18,19 Overexpression of H19 promotes proliferation and invasion of GC cells by actively binding to ISM1.20 HOTAIR participates in the drug resistance of GC cells through targeting miR-217.21 TET2 inhibits the growth of GC cells by regulating the expression of ANRIL.22 However, some key issues in systematic identification and characterization of candidate lncRNAs involved in GC, especially in the prognosis of GC patients, need to be resolved.
In the present study, we identified novel lncRNAs that were dysregulated in the development of GC. To comprehensively characterize those aberrantly expressed lncRNAs, we analyzed GC and normal tissue RNA sequencing data downloaded from The Cancer Genome Atlas (TCGA). In addition to identifying several GC-associated lncRNAs, we also discovered a novel lncRNA termed LINC00565. LINC00565 spans 2478 bp length and locates on chromosome 13, which is significantly upregulated in GC tissues. The elucidation of its functional association and underlying molecular mechanism involved in GC will provide better understanding of the role of lncRNA in gene regulation and tumor biology.
Methods
Data acquisition and analyses
Clinical data of 443 cases of GC and the corresponding genome-wide expression profiles were downloaded from the TCGA database (cancergenome.nih.gov). Data extracted from TCGA were standardized by the MD Anderson Cancer Center. The edger function was used to analyze differential genes, and the survival function was used for prognostic analysis. Gene Set Enrichment Analysis (GSEA) was performed using GSEA version 2.2.1 software. The downloaded microarray expression datasets, phenotype data file, and the MsigDB microarray platform annotation file were uploaded to the GSEA program. Enrichment analysis was performed according to the default weighted enrichment statistics method.
Sample collection
The paired GC tissues and the gastric mucosal tissues adjacent to GC tissues (>5 cm from the edge of the cancer nest) were derived from Danyang People’s Hospital of Jiangsu Province & Danyang Hospital affiliated to Nantong University. None of them received preoperative chemotherapy and/or radiotherapy. Resected samples were placed in a cryotube number and placed in a liquid nitrogen tank for long-term storage. Partial tissues were taken for clinical histopathological diagnosis. All patients signed an informed consent document for diagnosis and research on tissue specimens before being enrolled in the project. This research was approved by the ethics committee of Nantong University. All subjects gave written informed consent in accordance with the Declaration of Helsinki principles.
RNA extraction and qRT-PCR
Total RNA in GC tissues was extracted using TRIzol method for reverse transcription according to the instructions of PrimeScript RT reagent Kit (Takara, Tokyo, Japan). RNA concentration was detected using spectrometer. QRT-PCR was then performed based on the instructions of SYBR Premix Ex Taq TM (Takara). The amplification conditions were: Pre-denaturation at 94°C for 5 mins, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s. The relative gene expression was calculated using 2−△Ct method. Primers used in the study were as follows: GAPDH: F, 5′-CACCCACTCCTCCACCTTTG-3′, R, 5′-CCACCACCCTGTTGCTGTAG-3′; LINC00565: F, 5′-GGCCTGAGCATTGCATAACG-3′, R, 5′-CTGATGGAGCGACCGTCTAC-3′; AKT3: F, 5′-TGAAGTGGCACACACTCTAACT-3′, R, 5′-CCGCTCTCTCGACAAATGGA-3′; MicroRNA-665: F, 5′-GGTCTACAAAGGGAAGC-3′, R, 5′-TTTGGCACTAGCACATT-3′.
Cell culture and transfection
Human normal gastric epithelial cell line (GES-1) and GC cell lines (HGC, MGC, MKN28, MKN25, and AGS) were obtained from ATCC. Cells were cultured in DMEM containing 10% FBS (fetal bovine serum), 100 U/mL penicillin, and 100 μg/mL streptomycin (Hyclone, South Logan, UT, USA). Cells were incubated in a 5% CO2 incubator at 37°C.
For cell transfection, 1.5 mL of serum-free DMEM and 500 μL of mixture containing transfection reagent and LipofectamineTM 2000 were added in each well. After 4–6 hrs, complete medium was replaced. MicroRNA-665 mimics, microRNA-665 inhibitor, miR-NC, pcDNA-AKT3, and pcDNA-NC were constructed by GenePharma, Shanghai, China. Plasmid sequences were sh-LINC00565 1#: GCCTCCTGGTTTATAGCATGA; sh-LINC00565 2#: GGAGGACGCATCAATCCTTCT; sh-LINC00565 3#: GCTTTGCGAAGACTTTCTTCT; sh-LINC00565 4#: GAGCGAGATCCCATCCAAAC.
CCK-8
Transfected GC cells for 24 hrs were seeded into 96-well plates with 2×103 per well. Ten microliters of CCK-8 solution (cell counting kit-8, Dojindo, Kumamoto, Japan) was added in each well after cell culture for 2 days. The absorbance at 450 nm of each sample was measured by a microplate reader (Bio-Rad, Hercules, CA, USA).
Colony formation assay
GC cells in the logarithmic growth phase were gradiently seeded in the culture dish containing 10 mL of 37°C pre-warmed medium with 50, 100, and 200 cells per dish, respectively. After cell culture for 2–3 weeks until visible colonies formed, cells were washed with PBS and fixed with 4% paraformaldehyde. Colonies were stained with crystal violet for 10–30 mins and captured using the microscope.
Apoptotic determination
GC cells were prepared for suspension and adjusted to 1×106/mL. A total of 100 μL of suspension, 10 μL of Annexin-V, 380 μL of buffer, and 10 μL of PI were added in each tube. After 15-min incubation in dark, apoptotic rate was determined using flow cytometry.
Dual-luciferase reporter gene assay
GC cells were seeded in the 12-well plate at a density of 50%. Luciferase reporter plasmids were transfected until 70% of the cell density. According to sequences of AKT3 and LINC00565, we constructed AKT3 WT 3′UTR, AKT3 MT 3′UTR, LINC00565 WT, and LINC00565 MT. GC cells were co-transfected with 50 pmol/L microRNA-665 mimics/miR-NC and 80 ng AKT3 WT 3′UTR/AKT3 MT 3′UTR for 48 hrs. After cell wash with PBS, 50 μL of diluted 1×PLB was added for complete lysis. Luciferase activity was detected following the manufacturer’s recommendation of the commercial kit.
RNA-binding protein immunoprecipitation (RIP)
GC cells were washed and cross-linked with 0.01% formaldehyde for 15 mins. After centrifugation and cell lysis, extracted cells were incubated with RIP buffer containing protein A/G magnetic beads coated with anti-Ago2 or negative control anti-IgG antibody. After incubation at 4°C overnight, cells were incubated with Protein G-Sepharose for 3 hrs at 4°C, followed by the isolation of RNA. Target gene level was then detected by qRT-PCR.
Western blot
GC cells were lysed to harvest total cellular protein, followed by determination of total protein concentration. An equal amount of protein sample was loaded onto a 10% SDS-PAGE and then transferred to a polyvinylidene fluoride membrane. Membranes were blocked with skim milk and incubated with primary antibody (Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. On the other day, membranes were incubated with horseradish peroxidase conjugated secondary antibody for 2–3 hrs at room temperature. Finally, an image of the protein band was captured by the Tanon detection system using ECL reagent (Thermo, Waltham, MA, USA).
Statistical analyses
SPSS 22.0 statistical software was used for data analysis. Data were expressed as mean±standard deviation 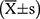 . Measurement data and classification data were compared using the student's t-test and chi-square test, respectively. Survival curves were introduced by Graphpad. P<0.05 considered the difference was statistically significant.
. Measurement data and classification data were compared using the student's t-test and chi-square test, respectively. Survival curves were introduced by Graphpad. P<0.05 considered the difference was statistically significant.
Results
LINC00565 was highly expressed in GC tissues and negatively correlated with the prognosis
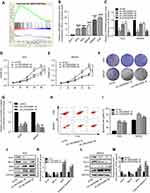
The clinical information and lncRNA expression profiles of GC patients were downloaded from the TCGA database for differential analyses (Figure 1A). We found that LINC00565 was remarkably upregulated in GC tissues (Figure 1B). In addition, Kaplan–Meier and log-rank tests were used to analyze the relationship between LINC00565 expression with disease free survival (DFS) and overall survival (OS) of GC patients. The results showed that high expression of LINC00565 was significantly negatively correlated with the DFS (Figure 1C) and OS in GC patients (Figure 1D). The higher the expression of LINC00565, the shorter the survival. Subsequently, we verified the expression of LINC00565 in GC patients admitted in our hospital. LINC00565 in GC tissues collected in our hospital was highly expressed, which was consistent with the results analyzed by the TCGA database (Figures 1E and S1A). By exploring the relationship between the expression level of LINC00565 and clinical data of GC, we found the expression of LINC00565 in GC tissue larger than 5 cm was significantly higher than those smaller than 5 cm (Figure 1F). Besides, GC patients with advanced stage showed higher expression of LINC00565 than those in early stage (Figure 1G). Similarly, the relationship between LINC00565 expression with DFS and OS of GC patients in our hospital was analyzed. High expression of LINC00565 was significantly negatively correlated with the DFS (Figure 1H) and OS (Figure 1I), which was consistent with the TCGA results. The above data indicated the potential role of LINC00565 in GC, which was negatively correlated to prognosis of GC patients.
LINC00565 knockdown inhibited proliferation and induced apoptosis of GC cells
GSEA analysis found that LINC00565 primarily regulates cell apoptosis (Figure 2A). To verify the in vitro functions of LINC00565, we examined the expression of LINC00565 in human normal gastric epithelial cell line and GC cell lines by qRT-PCR. LINC00565 was highly expressed in GC cell lines, especially in MKN25 and AGS cells (Figures 2B and S1B). Subsequently, LINC00565 interference sequences shRNA1#, shRNA2#, shRNA3#, and shRNA4# were transfected into MKN25 and AGS cells, respectively, and the expression of LINC00565 was detected. Transfection of shRNA1# and shRNA4# markedly downregulated the mRNA level of LINC00565, while shRNA2# and shRNA3# showed poor transfection efficacy (Figure 2C). Hence, shRNA1# and shRNA4# were selected for subsequent functional verification. After knockdown of LINC00565, CCK-8 assay was conducted to access cell proliferation. The data showed that proliferative rate of GC cells remarkably decreased after LINC00565 knockdown (Figure 2D and E). Colony formation assay revealed that LINC00565 knockdown inhibited colony formation ability of GC cells (Figure 2F and G). Apoptosis of GC cells was induced after LINC00565 knockdown (Figure 2H and I). Besides, expression levels Ki-67 and BCL2 were significantly downregulated by LINC00565 knockdown. Meanwhile, BAX and cleaved caspase-3 were upregulated after silence of LINC00565 (Figure 2J–M). These results indicated that interference with the expression of LINC00565 can inhibit proliferation and induce apoptosis of GC cells.
LINC00565 promoted GC progression by upregulating AKT3 as a ceRNA
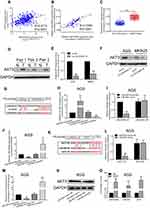
To further explore the mechanism of LINC00565 in GC development, we found that LINC00565 is positively correlated with the expression of AKT3 (Figure 3A). GC samples from our hospital further confirmed it (Figure 3B). The mRNA and protein levels of AKT3 remained high in GC tissues (Figure 3C and D), which were consistent with previous studies, indicating that AKT3 exerted as an oncogene.23,24 Afterward, MKN25 and AGS cells were transfected with shRNA-LINC00565 1#. Both mRNA and protein levels of AKT3 were downregulated after LINC00565 knockdown (Figure 3E and F).
Considering the mechanism of ceRNA,25,26 this study suggested that LINC00565 has the potential to bind to some miRNAs and prevent their degradation as a ceRNA. Through prediction in the Diana database (www.microrna.gr/LncBase), it is predicted that microRNA-665 could bind to LINC00565 (Figure 3G). Previous studies pointed out that microRNA-665 serves as a tumor-suppressor gene.27,28 Therefore, we chose microRNA-665 as a potential microRNA binding to LINC00565.
MicroRNA-665 mimics and microRNA-665 inhibitor were then constructed and their transfection efficacies in AGS cells were tested (Figure 3H). The binding condition between LINC00565 and microRNA-665 was verified. After constructing wild-type and mutant-type sequences (Figure 3G), dual-luciferase reporter gene was carried out. Luciferase activity decreased in LINC00565 WT 3′UTR group, whereas LINC00565 MUT 3′UTR did not show remarkable change, indicating LINC00565 can be combined with microRNA-665 to quench fluorescence (Figure 3I). Besides, it is found that microRNA-665 overexpression downregulated LINC00565 expression in GC cells and microRNA-665 knockdown obtained the opposite result (Figure 3J).
Through bioinformatics prediction, microRNA-665 could bind to AKT3 (Figure 3K). After constructing wild-type and mutant-type sequences (Figure 3K), luciferase activity decreased in AKT3 WT group, and AKT3 MT showed no obvious change (Figure 3L). By transfection of microRNA-665 mimics or microRNA-665 inhibitor in AGS cells, AKT3 expression was found to be negatively correlated to microRNA-665 expression at both mRNA and protein levels (Figure 3M and N). RIP results further confirmed the binding of LINC00565, microRNA-665, and AKT3 (Figure 3O).
AKT3 overexpression partially reversed the tumor suppressor role of LINC00565 in GC
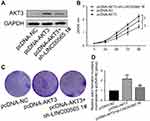
To verify whether AKT3 can reverse the effect of LINC00565 on GC development, we co-transfected with AKT3 overexpression plasmid and sh-LINC00565 in GC cells. AKT3 expression decreased in co-transfected cells compared with those only transfected with pcDNA-AKT3, suggesting that AKT3 could partially reverse the inhibitory effect of LINC00565 on its expression level (Figure 4A). CCK-8 and colony formation assay were conducted in co-transfected cells. Overexpression of AKT3 enhanced proliferative rate of GC cells, which was reduced after co-transfection, but was still higher than controls (Figure 4B). Similar results were obtained in colony formation assay (Figure 4C and D). These results indicated that overexpression of AKT3 partially reversed the anti-cancer effect of low expression of LINC00565 on GC development.
Discussion
GC is the fifth most common type of cancers worldwide, and it is an important public health problem.29 The high mortality rate of GC is associated with the lack of effective screening methods and the absence of apparent symptoms in early stage. Most GC patients have been in a malignant progression stage at the time of diagnosis. Moreover, the conventional chemotherapy and surgical treatment of GC have limited effects, leading to the poor prognosis of GC. Recently, epigenetic markers such as microRNAs and DNA methylation status have been shown to be involved in the development of GC.30 Therefore, it is highly desirable to find biomarkers that can be applied for the stable and effective diagnosis of GC.
In recent years, lncRNAs have been well concerned in tumor biology. Their abnormal regulation can lead to many human diseases including cancer.31 In this study, LINC00565 was found to be highly expressed in GC tissues through screening TCGA data, which was further verified in GC patients admitted in our hospital. Kaplan–Meier analysis was performed on the LINC00565 expression and the survival of GC patients. The data showed that the survival rate of GC patients with high expression of LINC00565 significantly shortened. The high expression of LINC00565 was associated with poor prognosis of GC. Hence, LINC00565 is expected to become a new prognostic marker for GC patients. Moreover, the low expression of LINC00565 can inhibit the proliferative and colony formation abilities of GC cells, but induce cell apoptosis. We speculated that LINC00565 may participate in the occurrence and development of GC.
LncRNAs can act as ceRNAs to adsorb microRNAs and promote the abnormal expressions of certain specific mRNAs. As a result, downregulation of a specific ceRNAs can promote the binding of more specific miRNAs to the corresponding mRNAs, thus reducing the expression levels of certain tumor-suppressor genes.25,26 Although the mechanism of ceRNAs in tumorigenesis is not fully understood, previous studies suggested that ceRNAs are complementary in post-transcriptional regulation of lncRNAs in GC. It is found that lncRNA-FER1L4 contains miRNA-binding elements and competitively binds to miR-106a-5p, thereby regulating the expression levels of different genes, such as PTEN, RB1, RUNX1, VEGFA, CDKN1A, E2F1, HIPK3, IL10, and PAK7. Subsequently, proliferation and transformation from G0/G1 phase to S phase of tumor cells are regulated by lncRNA-FER1L4.32 Studies in other types of tumors have also shown that lncRNAs exert the function of molecular sponges as ceRNAs.33–35 This study showed that LINC00565 and AKT3 could bind to microRNA-665.
MicroRNA-665 exerts a regulatory role in the human body. In ovarian cancer tissues, microRNA-665 inhibits proliferation and migration of cancer cells by regulating HOXA10.27 LINC00462 is involved in the development of pancreatic cancer by regulating microRNA-665.28 In this study, microRNA-655 bound to LINC00565 and AKT3, thereafter participating in the development of GC.
AKT3 is involved in cell proliferation, differentiation, apoptosis, and tumorigenesis. In gliomas, AKT3 promotes the progression of glioma cells.23 Decreased invasiveness is observed in T98G cells after knockdown of AKT3.24 Targeting on AKT3 can be an effective method for inhibiting the growth of breast cancer.36 In the present study, the expression of AKT3 significantly increased in GC tissues, which was positively correlated with the expression of LINC00565. Overexpression of AKT3 partially reversed the inhibitory effect of LINC00565 on GC development.
This study investigated the possible role of LINC00565 in the pathogenesis of GC and found that LINC00565 was abnormally expressed in GC tissues. Low expression of LINC00565 inhibited proliferative and migratory abilities of GC cells. Bioinformatics predicted that microRNA-665 bound to LINC00565 and AKT3. It is demonstrated that LINC00565 may act as a microRNA-665 sponge to positively regulate the expression of its target gene AKT3, thus regulating proliferation and migration of GC cells.
This study explored the specific role of LINC00565 in GC for the first time. Our results proposed that the axis of LINC00565/microRNA-665/AKT3 may be a promising target on diagnosis, treatment, and prognosis of GC.
There were still some limitations, however, that should be focused on in further studies. First of all, the biological role of microRNA-665 in GC was less verified. Whether microRNA-665 could reverse the functions of LINC00565 in GC development remains unknown. Second, we only investigated proliferative and colony formation abilities of GC cells. Other biological performances of GC cells, such as migration and invasiveness, are needed to be explored if they could be influenced by LINC00565 as well. Last but not least, animal experiments are required to explore the in vivo effect of LINC00565 on GC development.
Conclusion
LINC00565 is highly expressed in GC, which regulates proliferative and apoptotic capacities of GC cells by targeting microRNA-665 that binds to AKT3.
Acknowledgments
This work was supported by National Natural Science Foundation of China (31700444), Health Research Project of Jiangsu Province and Social Development Guide Project of Zhenjiang (FZ2015074).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
2. Taniguchi H, Moriya C, Igarashi H, et al. Cancer stem cells in human gastrointestinal cancer. Cancer Sci. 2016;107:1556–1562. doi:10.1111/cas.13069
3. Katuri V, Tang Y, Marshall B, et al. Inactivation of ELF/TGF-beta signaling in human gastrointestinal cancer. Oncogene. 2005;24:8012–8024. doi:10.1038/sj.onc.1208946
4. Noh SH, Park SR, Yang HK, et al. Adjuvant capecitabine plus oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): 5-Year follow-up of an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:1389–1396. doi:10.1016/S1470-2045(14)70473-5
5. Zeng ZM, Luo FF, Zou LX, et al. Human papillomavirus as a potential risk factor for gastric cancer: a meta-analysis of 1,917 cases. Onco Targets Ther. 2016;9:7105–7114. doi:10.2147/OTT.S115053
6. Berger H, Marques MS, Zietlow R, Meyer TF, Machado JC, Figueiredo C. Gastric cancer pathogenesis. Helicobacter. 2016;21(Suppl 1):34–38. doi:10.1111/hel.12338
7. Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi:10.1038/embor.2009.181
8. Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–18102. doi:10.1073/pnas.0709193104
9. Li X, Montgomery SB. Detection and impact of rare regulatory variants in human disease. Front Genet. 2013;4:67. doi:10.3389/fgene.2013.00067
10. Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. doi:10.1042/BST20120020
11. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi:10.1038/nrg3074
12. Li X, Wu Z, Fu X, Han W. LncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1–21. doi:10.1016/j.mrrev.2014.04.002
13. Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi:10.1093/nar/gks296
14. Mouraviev V, Lee B, Patel V, et al. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016;19:14–20. doi:10.1038/pcan.2015.48
15. Ellis BC, Molloy PL, Graham LD. CRNDE: a long non-coding RNA involved in cancer, neurobiology, and development. Front Genet. 2012;3:270. doi:10.3389/fgene.2012.00270
16. Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi:10.1002/jcb.24055
17. Yildirim E, Kirby JE, Brown DE, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi:10.1016/j.cell.2013.01.034
18. Wang J, Sun J, Wang J, et al. Long noncoding RNAs in gastric cancer: functions and clinical applications. Onco Targets Ther. 2016;9:681–697. doi:10.2147/OTT.S95412
19. Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142:2231–2237. doi:10.1007/s00432-016-2183-7
20. Jing W, Zhu M, Zhang XW, et al. The significance of long noncoding RNA h19 in predicting progression and metastasis of cancers: a meta-analysis. Biomed Res Int. 2016;2016:5902678. doi:10.1155/2016/5902678
21. Wang H, Qin R, Guan A, et al. HOTAIR enhanced paclitaxel and doxorubicin resistance in gastric cancer cells partly through inhibiting miR-217 expression. J Cell Biochem. 2018;119:7226–7234.
22. Deng W, Wang J, Zhang J, Cai J, Bai Z, Zhang Z. TET2 regulates LncRNA-ANRIL expression and inhibits the growth of human gastric cancer cells. IUBMB Life. 2016;68:355–364. doi:10.1002/iub.1490
23. Turner KM, Sun Y, Ji P, et al. Genomically amplified Akt3 activates DNA repair pathway and promotes glioma progression. Proc Natl Acad Sci U S A. 2015;112:3421–3426. doi:10.1073/pnas.1414573112
24. Paul-Samojedny M, Pudelko A, Kowalczyk M, et al. Knockdown of AKT3 and PI3KCA by RNA interference changes the expression of the genes that are related to apoptosis and autophagy in T98G glioblastoma multiforme cells. Pharmacol Rep. 2015;67:1115–1123. doi:10.1016/j.pharep.2015.04.012
25. Li D, Yang M, Liao A, et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J Cell Mol Med. 2018;22(8):3875–3886.
26. Xu Y, Zhang G, Zou C, et al. LncRNA MT1JP suppresses gastric cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem. 2018;46:2445–2459. doi:10.1159/000489651
27. Liu J, Jiang Y, Wan Y, Zhou S, Thapa S, Cheng W. MicroRNA665 suppresses the growth and migration of ovarian cancer cells by targeting HOXA10. Mol Med Rep. 2018;18:2661–2668. doi:10.3892/mmr.2018.9252
28. Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 promotes pancreatic cancer invasiveness through the microRNA-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis. 2018;9:706. doi:10.1038/s41419-018-1111-y
29. Shipley WU, Seiferheld W, Lukka HR, et al. Radiation with or without antiandrogen therapy in recurrent prostate cancer. N Engl J Med. 2017;376:417–428. doi:10.1056/NEJMoa1607529
30. Zhao J, Nie Y, Wang H, Lin Y. MiR-181a suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Gene. 2016;576:828–833. doi:10.1016/j.gene.2015.11.013
31. Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–157. doi:10.1038/nrm.2017.104
32. Xia T, Liao Q, Jiang X, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi:10.1038/srep06088
33. Li D, Yang M, Liao A. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J Cell Mol Med. 2018;22:8. doi:10.1111/jcmm.13661
34. Zhang G, Li S, Lu J, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17(1):87. doi:10.1186/s12943-018-0829-6
35. H Y T, Wang C, Liu G, et al. Long noncoding RNA NEAT1‐modualted miR‐506 regulates gastric cancer development through targeting STAT3. J Cell Biochem. 2018;120:4.
36. Chin YR, Yoshida T, Marusyk A, Beck AH, Polyak K, Toker A. Targeting Akt3 signaling in triple-negative breast cancer. Cancer Res. 2014;74:964–973. doi:10.1158/0008-5472.CAN-13-2175
Supplementary material
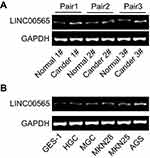 |
Figure S1 LINC00565 was upregulated in gastric cancer tissues (A) and cell lines (B). |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.