Back to Journals » OncoTargets and Therapy » Volume 12
Linc00511 Indicates A Poor Prognosis Of Liver Hepatocellular Carcinoma
Authors Hu P, Cui H, Lei T, Li S, Mai E, Jia F
Received 22 August 2019
Accepted for publication 18 October 2019
Published 8 November 2019 Volume 2019:12 Pages 9367—9376
DOI https://doi.org/10.2147/OTT.S228231
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Pingan Hu,1 Huxiao Cui,2 Ting Lei,1 Siqiao Li,1 Erhui Mai,1 Fuxin Jia1
1Department of Hepatobiliary Surgery, Luoyang Center Hospital, Luoyang, People’s Republic of China; 2Department of Hepatobiliary Surgery, Xuchang Central Hospital, Xuchang, People’s Republic of China
Correspondence: Pingan Hu
Department of Hepatobiliary Surgery, Luoyang Center Hospital, No. 288, Zhongzhou Middle Road, Xigong District, Luoyang, Henan 471000, People’s Republic of China
Tel +86379-63892085
Email [email protected]
Objective: To uncover the specific function of linc00511 in the progression of liver hepatocellular carcinoma (LIHC) and the underlying mechanism.
Patients and methods: GEPIA dataset containing 9736 LIHC samples and 857 normal samples were downloaded from TCGA. Expression pattern and prognostic potential of linc00511 in LIHC were analyzed. Subsequently, expression level of linc00511 in LIHC tissues collected in our hospital and cell lines were determined by quantitative real-time polymerase chain reaction (qRT-PCR). Differential expressions of linc00511 in LIHC with different tumor grades and metastatic status were compared. After transfection of si-linc00511, proliferative and migratory changes in Huh7 and Hep3B cells were assessed by cell counting kit-8 (CCK-8), 5-ethynyl-2ʹ-deoxyuridine (EdU) and Transwell assay. Lastly, Pearson correlation analysis and qRT-PCR were conducted to investigate the interaction between linc00511 and miR-29c.
Results: Linc00511 was upregulated in LIHC tissues and cell lines. Its level was positively correlated to TNM staging, lymphatic metastasis and poor prognosis in LIHC patients. Knockdown of linc00511 attenuated proliferative and migratory abilities in Huh7 and Hep3B cells. In addition, miR-29c was downregulated in LIHC and negatively linked to linc00511 level. A negative interaction between linc00511 and miR-29c could be a regulatory feedback influencing the progression of LIHC.
Conclusion: Linc00511 accelerates the proliferation and migration in LIHC, thus aggravating tumor progression. Meanwhile, linc00511 could be utilized as a hallmark predicting poor prognosis in LIHC patients.
Keywords: LIHC, linc00511, miRNA-29c, proliferation, migration
Introduction
Over the past few years, more than 700,000 people die of liver hepatocellular carcinoma (LIHC) each year, and this number has increased annually.1 Symptoms and signs of early-stage LIHC are atypical. Most of LIHC cases are initially diagnosed in advanced stage with worse disease condition and higher malignant level.2 Compared with early primary LIHC, distant metastasis in advanced stage is the major reason for triggering tumor death.3 LIHC is a complex tumor disease with strong invasiveness. Its pathology includes changes in tumor cell behaviors, vascular system, etc.4 It is urgent to uncover new effective biomarkers for LIHC, so as to improve the clinical outcome.
Long non-coding RNA (lncRNA) is an autonomous transcribed RNA that does not encode a protein. Its minimum length is about 200 nucleotides.5 With the development of high-throughput sequencing technology, current research data showed that 90% RNAs in the human genome belong to non-coding RNAs. Non-coding RNAs exert crucial functions in regulating cellular behaviors and tumor diseases through influencing tumor cells and intracellular signals.6,7 For example, linc00460 stimulates the progression of head and neck tumors through promoting apoptosis and autophagy by targeting miR-206.8 LncRNA RP1 is considered as an important oncogene in breast cancer through suppressing the p27kip1 pathway.9 In liver cancer, lncRNA TPTEP1 enhances cisplatin-sensitivity by regulating the phosphorylation of STAT3.10 A previous study has uncovered that linc00511 is a potential oncogene. Silence of linc00511 in breast cancer enhances the therapeutic efficacy of paclitaxel-induced chemotherapy.11 Besides, the carcinogenic role of linc00511 has also been confirmed in ovarian cancer and NSCLC.12,13 Through analyzing the dataset downloaded from TCGA, it is identified that linc00511 was abnormally expressed in LIHC, which was consistent with previously reported.14 Our further experiments uncovered a potential regulatory network between linc00511 and other target miRNAs. Their interaction may be a novel mechanism of LIHC development.
Patients And Methods
TCGA Dataset Analyses
Linc00511 expression and its prognostic potential in LIHC were assessed by analyzing dataset downloaded from GEPIA (http://GEPIA.cancer-pku.cn/index.html).15 The GEPIA online database contains research data both from TCGA and GTEx, involving sequencing data of 9736 tumor samples and 857 normal samples.
Sample Collection
Forty LIHC tissues and matched paracancerous tissues were surgically resected in Luoyang Center Hospital from May 2016 to December 2018. All samples were pathologically confirmed. Patients and their families in this study have been fully informed. All patients provided written informed consents. This study was conducted in accordance with the Declaration of Helsinki. This study was approved by Ethics Committee of Luoyang Center Hospital.
Cell Culture
Normal hepatocytes (LO2) and liver cancer cell lines (MHCC-97H, Huh7, HCC-LM3, Hep3B, MHCC-97L and Huh6) were purchased from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle's medium (DMEM) (Gibco, Rockville, MD, USA) containing 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA), and 1% penicillin–streptomycin in a 5% CO2 at 37°C. Cell passage should be limited within 30 generations.
Transfection
Cells in good growth condition were transfected with si-linc00511, pcDNA-linc00511, miRNA-29c mimics or negative control (Riobobio, Guangzhou, China) using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Transfected cells for 24–48 hrs were harvested for the following experiments.
RNA Extraction And Quantitative Real-Time Polymerase Chain Reaction (QRT-PCR)
Cells were lysed to harvest RNAs using TRIzol method (Invitrogen, Carlsbad, CA, USA), and the extracted RNAs were subjected to reverse transcription according to the instructions of PrimeScript RT reagent Kit (TaKaRa, Tokyo, Japan). RNA concentration was detected using a spectrometer. QRT-PCR was then performed based on the instructions of SYBR Premix Ex Taq TM (TaKaRa, Tokyo, Japan). Relative level was calculated using 2−ΔΔCt method. Linc00511: Forward: 5ʹ-AGGGGCGACTACTGTTACCT-3ʹ and reverse: 5ʹ-CGTCCAAACAGGCTGGATCT-3′; GAPDH: Forward: 5ʹ-AACTTTGGCATTGTGGAAGG-3ʹ and reverse: 5ʹ-CACATTGGGGGTAGGAACAC-3′; MiRNA-29c: Forward: 5ʹ-GCGCGCGTAGCACCATTTGAAAT-3ʹ and reverse: 5ʹ-ATCCAGTGCAGGGTCCGAGG-3ʹ; U6: Forward: 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ and reverse: 5ʹ-GGAACGCTTCACGAATTTG-3′.
Cell Counting Kit-8 (CCK-8) Assay
Cells were seeded into 96-well plates with 5×103 cells per well. At the appointed time points, 10 μL of CCK-8 solution (Dojindo, Kumamoto, Japan) was added in each well. The absorbance at 450 nm of each sample was measured by a microplate reader (Bio-Rad, Hercules, CA, USA).
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
Cells were inoculated into 96-well plates with 1×105 cells per well, and labeled with 100 μL of EdU reagent (50 μM) per well for 2 hrs (Solarbio, Beijing, China). After washing with phosphate-buffered saline (PBS), cells were fixed in 50 μL of fixation buffer, decolored with 2 mg/mL glycine and permeated with 100 μL of penetrant. After washing with PBS once, cells were stained with Hochest in dark for 30 mins. EdU-positive ratio was determined under a fluorescent microscope (magnification 20×).
Wound Healing Assay
Wound healing assay was conducted to assess cell migratory ability. Transfected cells were cultured to about 80% confluence and an artificial wound on cell plate was created using a sterile pipette tip. Wound healing was observed at 0 and 24 hrs using an inverted optical microscope (Nikon, Japan).
Transwell
Cell density was adjusted to 3×104 cells/mL. 100 μL of suspension was applied in the upper side of Transwell chamber (Corning, Corning, NY, USA) coated with diluted Matrigel. In the bottom side, 600 μL of medium containing 20% FBS was applied. After 48 hrs of incubation, cells penetrated to the bottom side were fixed in 4% paraformaldehyde for 20 mins, stained with crystal violet for 20 mins and counted using a microscope. The number of penetrating cells was counted in 5 randomly selected fields per sample (magnification 200×).
Statistical Analysis
Graphpad prism 7.0 (La Jolla, CA, USA) was used for data analysis. Data were expressed as mean ± standard deviation (x̅±SD). Pearson correlation test was conducted to compare the relationship between two genes. Survival analysis was performed by introducing Kaplan–Meier method. Intergroup data were compared using the t-test. P<0.05 considered the difference was statistically significant.
Results
Linc00511 Was Upregulated In LIHC And Related To Poor Prognosis
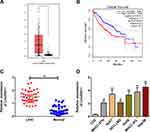
Through analyzing TCGA dataset, linc00511 was found to be upregulated in LIHC tissues compared with normal liver tissues (Figure 1A). Survival analysis showed that high level of linc00511 in LIHC patients predicted worse prognosis (Figure 1B). Subsequently, we determined linc00511 in LIHC and matched paracancerous tissues collected in our hospital. Identically, linc00511 was highly expressed in LIHC tissues relative to normal ones (Figure 1C). In vitro expression of linc00511 was similarly upregulated in liver cancer cells (Figure 1D). The above data demonstrated a potential carcinogenic role of linc00511 in LIHC.
Linc00511 Was Linked To Lymphatic Metastasis And Tumor Staging Of LIHC
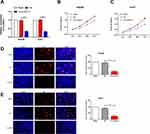
According to the collected clinical data, LIHC patients were classified based on their tumor staging and statue of lymphatic metastasis. In LIHC patients with advanced stages (III–IV), linc00511 level was higher than those in stages I–II (Figure 2A). Expression level of linc00511 remained higher in LIHC patients with lymphatic metastasis relative to those with negative metastasis (Figure 2B).
Knockdown Of Linc00511 Attenuated Proliferative And Migratory Abilities Of LIHC
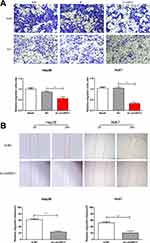
Hep3B and Huh7 cells were selected for the following in vitro experiments. First of all, transfection efficacy of si-linc00511 was verified in Hep3B and Huh7 cells (Figure 3A). Transfection of si-linc00511 greatly decreased viability in liver cancer cells as CCK-8 assay revealed (Figure 3B and C). Besides, EdU assay uncovered a decline in EdU-positive ratio after knockdown of linc00511 in Hep3B and Huh7 cells, further verifying an inhibited proliferative ability (Figure 3D and E). Regulatory effect of linc00511 on migratory ability of LIHC was assessed through Transwell assay and wound healing assay. Transfection of si-linc00511 remarkably reduced migratory cell number, confirming the attenuated migratory ability in LIHC, evidenced by results of Transwell and wound healing assay (Figure 4A and B).
A Negative Interaction Between miRNA-29c And Linc00511 In LIHC
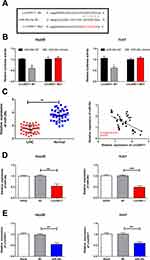
Previous study has demonstrated that miRNA-29c is a potential target of linc00511.16 Dual-luciferase reporter gene assay was conducted to assess the binding relationship between linc00511 and LIHC. Decreased luciferase activity in wild-type linc00511 vector overexpressing miRNA-29c, confirming that miRNA-29c was the target binding linc00511 (Figure 5A and B). In this experiment, miRNA-29c was downregulated in LIHC tissues relative to controls (Figure 5C). Pearson correlation test uncovered a negative correlation between linc00511 and miRNA-29c levels in LIHC tissues (Figure 5C). Moreover, linc00511 overexpression markedly downregulated miRNA-29c level in Hep3B and Huh7 cells (Figure 5D). Similarly, linc00511 level was downregulated by overexpression of miRNA-29c in liver cancer cells (Figure 5E). Thus, a negative interaction between linc00511 and miRNA-29c in LIHC was clarified. MiRNA-29c may be a key gene involved in the malignant progression of LIHC regulated by linc00511.
Overexpression Of miRNA-29c Abolished Regulatory Effects Of Linc00511 On Migratory Abilities In LIHC
Linc00511 was closely correlated to lymphatic metastasis in LIHC patients. Besides, survival analysis uncovered that high level of linc00511 was linked to a poor prognosis in LIHC. In addition, migratory ability of LIHC cells was influenced by linc00511. Previous studies have demonstrated that miRNA-29c serves as a tumor-suppressor gene and is closely related to LIHC migration.17,18 It is speculated that miRNA-29c could be significant during the migratory process of LIHC. Our findings showed that overexpression of miRNA-29c partially reversed the inhibitory effects of linc00511 on the migration of LIHC, as transwell (Figure 6A and B) and wound healing assay results (Figure 6C and D) revealed. Therefore, miRNA-29c was responsible for the linc00511-regulated progression of LIHC.
Discussion
For a long period, lncRNAs are considered as non-functional products formed during the transcription progression of RNA polymerases.19 With the advance in medical technologies, lncRNAs are found to be important in physiological and pathological processes, which have been well concerned.20 It is believed that lncRNAs are key factors in regulating tumor microenvironment and therapeutic targets for tumor treatment.21 LIHC is a malignancy with high recurrence and mortality. Diagnosis and intervention as early as possible are the most effective approaches to reduce tumor-related deaths. It is reported that lncRNA SNHG10 leads to poor prognosis of liver cancer through aggravating the malignant level and invasive ability.22 LncRNA CRNDE accelerates liver cancer progression via regulating the mRNA level of SIX1.23
Through analyzing the TCGA dataset, linc00511 was upregulated in LIHC tissues and closely related to poor prognosis. It is suggested that linc00511 may be an oncogene in LIHC, which was consistent with previous conclusions.13,24 Moreover, our findings showed that linc00511 was correlated to tumor staging and lymphatic metastasis of LIHC. Through a series of functional experiments, linc00511 was proved to promote proliferative and migratory abilities of LIHC.
Through bioinformatics prediction and literature review, miRNA-29c could be a potential target binding linc00511, which was further confirmed in our experiments. As a tumor-suppressor gene, miRNA-29c is able to suppress tumor cell growth and proliferation through regulating tumor-related genes. It is found to be related to AFP level in liver cancer patients.17,25–27 Furthermore, we analyzed miRNA-29c level in LIHC samples, which was downregulated in tumor tissues relative to paracancerous ones. Pearson correlation test identified a negative relationship between levels of linc00511 and miRNA-29c in LIHC tissues. In addition, a negative interaction between linc00511 and miRNA-29c was verified in Huh7 and Hep3B cells. MiRNA-29c may be responsible for linc00511-regulated progression of LIHC.
Conclusion
Linc00511 aggravates the progression of LIHC through accelerating proliferative and migratory abilities of tumor cells. Linc00511 may serve as a prognostic biomarker for LIHC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gupta M, Gabriel H, Miller FH. Role of imaging in surveillance and diagnosis of hepatocellular carcinoma. Gastroenterol Clin North Am. 2018;47(3):585–602. doi:10.1016/j.gtc.2018.04.013
2. Si T, Chen Y, Ma D, et al. Transarterial chemoembolization prior to liver transplantation for patients with hepatocellular carcinoma: a meta-analysis. J Gastroenterol Hepatol. 2017;32(7):1286–1294. doi:10.1111/jgh.13727
3. Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58(1):205–217. doi:10.1002/hep.26315
4. Dufour JF, Johnson P. Liver cancer: from molecular pathogenesis to new therapies: summary of the EASL single topic conference. J Hepatol. 2010;52(2):296–304. doi:10.1016/j.jhep.2009.11.010
5. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi:10.1016/j.cell.2014.03.008
6. Wang C, Jing J, Cheng L. Emerging roles of non-coding RNAs in the pathogenesis, diagnosis and prognosis of osteosarcoma. Invest New Drugs. 2018;36(6):1116–1132. doi:10.1007/s10637-018-0624-7
7. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
8. Xue K, Li J, Nan S, Zhao X, Xu C. Downregulation of LINC00460 decreases STC2 and promotes autophagy of head and neck squamous cell carcinoma by up-regulating microRNA-206. Life Sci. 2019;231:116459. doi:10.1016/j.lfs.2019.05.015
9. Jia X, Shi L, Wang X, et al. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation. Cell Death Dis. 2019;10(5):373. doi:10.1038/s41419-019-1566-5
10. Ding H, Liu J, Zou R, Cheng P, Su Y. Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma progression by suppressing STAT3 phosphorylation. J Exp Clin Cancer Res. 2019;38(1):189. doi:10.1186/s13046-019-1193-0
11. Zhang H, Zhao B, Wang X, Zhang F, Yu W. LINC00511 knockdown enhances paclitaxel cytotoxicity in breast cancer via regulating miR-29c/CDK6 axis. Life Sci. 2019;228:135–144. doi:10.1016/j.lfs.2019.04.063
12. Wang J, Tian Y, Zheng H, Ding Y, Wang X. An integrated analysis reveals the oncogenic function of lncRNA LINC00511 in human ovarian cancer. Cancer Med. 2019;8(6):3026–3035. doi:10.1002/cam4.2171
13. Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids. 2016;5(11):e385. doi:10.1038/mtna.2016.94
14. Lim LJ, Wong S, Huang F, et al. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 2019;79:5131–5139. doi:10.1158/0008-5472.CAN-19-0255
15. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
16. Wang W, Lou W, Ding B, et al. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY). 2019;11(9):2610–2627. doi:10.18632/aging.101933
17. Wu H, Zhang W, Wu Z, et al. miR-29c-3p regulates DNMT3B and LATS1 methylation to inhibit tumor progression in hepatocellular carcinoma. Cell Death Dis. 2019;10(2):48. doi:10.1038/s41419-018-1281-7
18. Bae HJ, Noh JH, Kim JK, et al. MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene. 2014;33(20):2557–2567. doi:10.1038/onc.2013.216
19. Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37(4):144–151. doi:10.1016/j.tibs.2011.12.003
20. Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474(24):4219–4251. doi:10.1042/BCJ20170079
21. Patel JS, Hu M, Sinha G, et al. Non-coding RNA as mediators in microenvironment-breast cancer cell communication. Cancer Lett. 2016;380(1):289–295. doi:10.1016/j.canlet.2015.11.016
22. Lan T, Yuan K, Yan X, et al. LncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res. 2019;79(13):3220–3234. doi:10.1158/0008-5472.CAN-18-4044
23. Tang D, Zhao L, Peng C, Ran K, Mu R, Ao Y. LncRNA CRNDE promotes hepatocellular carcinoma progression by upregulating SIX1 through modulating miR-337-3p. J Cell Biochem. 2019;120:16128–16142. doi:10.1002/jcb.28894
24. Yang F, Lyu S, Dong S, Liu Y, Zhang X, Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther. 2016;9:761–772. doi:10.2147/OTT.S97664
25. Ji J, Chen H, Liu XP, et al. A miRNA combination as promising biomarker for hepatocellular carcinoma diagnosis: a study based on bioinformatics analysis. J Cancer. 2018;9(19):3435–3446. doi:10.7150/jca.26101
26. Li W, Yi J, Zheng X, et al. miR-29c plays a suppressive role in breast cancer by targeting the TIMP3/STAT1/FOXO1 pathway. Clin Epigenetics. 2018;10:64. doi:10.1186/s13148-018-0495-y
27. Wang L, Yu T, Li W, et al. The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3. Oncogene. 2019;38(17):3134–3150. doi:10.1038/s41388-018-0642-0
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.