Back to Journals » OncoTargets and Therapy » Volume 12
LINC00491 as a new molecular marker can promote the proliferation, migration and invasion of colon adenocarcinoma cells
Authors Wan J , Deng D, Wang X , Wang X, Jiang S , Cui R
Received 11 January 2019
Accepted for publication 8 June 2019
Published 13 August 2019 Volume 2019:12 Pages 6471—6480
DOI https://doi.org/10.2147/OTT.S201233
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Jiahui Wan,1,2 Daiqian Deng,3 Xiuli Wang,1,4 Xiaojin Wang,1 Shijun Jiang,1,5 Rongjun Cui1
1Department of Biochemistry and Molecular Biology, Mudanjiang Medical University, Mudanjiang, Heilongjiang, People’s Republic of China; 2Department of Clinical Laboratory, Harbin Public Security Hospital, Harbin, Heilongjiang, People’s Republic of China; 3Department of Biology, Mudanjiang Medical University, Mudanjiang, Heilongjiang, People’s Republic of China; 4Department of Clinical Laboratory, The Seventh Hospital in Qiqihar, Qiqihar, Heilongjiang, People’s Republic of China; 5Department of Clinical Laboratory, Daqing Medical College, Daqing, Heilongjiang, People’s Republic of China
Background: Long non-coding RNAs (lncRNAs) play an important role in the pathogenesis of multiple tumors. However, the roles of lncRNAs during colon adenocarcinoma and cancer progression remain unclear. This study aimed identify new lncRNAs that act as molecular markers for the prevention and diagnosis of colon adenocarcinoma.
Methods: RNA sequencing (RNA-Seq) data associated with colon adenocarcinoma were retrieved from the Cancer Genome Atlas (TCGA). Biological processes in Gene Ontology (Go) and the Kyoto Encyclopedia of Genomes (KEGG) were searched for pathways at the significance level. The expression of LINC00491 and its downstream targets were assessed by real-time PCR, Western blotting and dual-luciferase assays. Biological functions of LINC00491 during cell proliferation, migration and invasion were assessed using CCK-8, colony formation assays, wound healing, and transwell invasion assays in colon adenocarcinoma HT-29 and HCT116 cells.
Results: Bioinformatics analysis with the TCGA colon adenocarcinoma dataset showed that LINC00491 was significantly up-regulated in colon adenocarcinoma. Furthermore, we found that LINC00491 positively regulates SERPINE1 expression through sponging miR-145 and promoting the proliferation, migration, and invasion of colon adenocarcinoma cells, thus playing an oncogenic role during colon adenocarcinoma pathogenesis.
Conclusion: LINC00491 functions as a ceRNA to promote SERPINE1 expression by sponging miR-145. LINC00491 serves as a therapeutic target and prognostic biomarker in colon adenocarcinoma.
Keywords: LINC00491, colon adenocarcinoma, competing endogenous RNA, cell proliferation, migration, invasion
Introduction
Colon adenocarcinoma is the third leading cause of cancer-related death in the United States.1 In China, colon adenocarcinoma is the most common gastrointestinal malignancy.2 According to the 2018 Global Cancer statistics report, new cases of colon adenocarcinoma accounted for 6.1% of all cancer cases, and the death toll accounted for 5.8% of all cases.3 As patients are asymptomatic during the early stages, colon adenocarcinomas are diagnosed at an advanced stage with malignant proliferation and metastasis. Although the clinical treatment of colon adenocarcinoma has improved, including surgical techniques, radiotherapy, and chemotherapy, the prognosis of patients diagnosed with colon adenocarcinoma remains unsatisfactory. Consequently, the discovery of novel regulatory factors and therapeutic targets for colon adenocarcinoma are required to improve our understanding of cancer occurrence and disease progression.
LncRNAs are ≥200 nucleotides in length and regulate an array of cell biological processes including chromatin remodeling, transcription, and post-transcriptional events.4,5 The most recognized molecular mechanism of lncRNAs are to act as miRNA “sponges” to regulate downstream target genes.6,7 LncRNAs act as competing endogenous RNAs (ceRNAs) to accelerate cancer proliferation, migration, and invasion. The E2F-mediated MNX-AS1-miR-218-5p-SEC61A1 feedback loop contributes to the progression of colon adenocarcinoma.8 LncRNA CCAT2 plays an oncogenic role in Cholangiocarcinoma (CCA) and CCAT2 knockdown inhibits the migration and invasion of CCA cells.9 HOXD-AS1 functions as a ceRNA that competitively binds to miR-130a-3p and up-regulates SOX4. Promoting hepatocellular carcinoma cell metastasis.10 LncRNA-HNF1A-AS1 regulates the proliferation and migration of oesophageal adenocarcinoma cells.11
Long intergenic non-protein coding RNA 491 (LINC00491) is located on chromosome 5q21.1 in the human genome and is significantly up-regulated in colon adenocarcinoma tissue. This study revealed that LINC00491 promotes colon adenocarcinoma cell proliferation, migration and invasion by competitively binding to miR-145 and up-regulating SERPINE1. LINC00491 therefore not only acts as a prognostic biomarker but as a therapeutic target for colon adenocarcinoma.
Materials and methods
Patients and samples from the TCGA database
To search for specific lncRNAs, miRNAs and mRNAs in colon adenocarcinoma, we downloaded gene expression data from the TCGA database (https://portal.gdc.cancer.gov/, up to April 9, 2018, including 480 tumors and 41 adjacent non-tumor tissues).
RNA sequence data processing and differential expression analysis
Differentially expressed mRNAs (DEmRNAs), differentially expressed lncRNAs (DElncRNAs) and differentially expressed miRNAs (DEmiRNAs) were identified using the EdgeR package in R (version 3.4.1) between colon adenocarcinoma and adjacent-normal colon tissue. Adjusted p-values<0.01 and |logFC|>2 were set as the cut-off criteria. Ggplot2 packages in R were used to visualize volcano plots and heat maps.
Establishment of the ceRNA network
The miRcode online tool (http://www.mircode.org) were used to predict DElncRNAs-DEmiRNAs interactions in colon adenocarcinoma. miRDB (http://www.mirdb.org), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), and Targetscan (http://www.targetscan.org) databases were used to predict the DEmRNAs targeted by DEmiRNAs. Finally, ceRNA networks were established by DEmiRNAs that regulate the expression of DEmRNAs and DElncRNAs, DElncRNAs-DEmiRNAs-DEmRNAs ceRNA networks were visualized using Cytoscape (version 3.5.1).
Functional enrichment analysis
GO enrichment analysis were performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/). FDR<0.05 were set as the cut-off criterion. KEGG pathway analysis was performed with packages in R. The threshold was p<0.05. The R clusterProfiler package was used to predict potential functions for the DEmRNAs in the ceRNA network.
Human colon adenocarcinoma cell complementary DNA (cDNA)
LINC00491 expression in human colon adenocarcinoma cell lines was detected by human colon adenocarcinoma cell complementary DNA (cDNA) chip 36 points (OUTDO BIOTECH, Shanghai, China). cDNA was derived from human colon adenocarcinoma cells (T84, RKO, COLO-205, LOVO, COLO320DM, COLO320, SW480, HCT-15, SW620, HT-29, HCT116) and normal colon epithelial cell lines (NCM460) with 25 ng of cDNA templates per well. Experiments were performed in triplicate. Gene expression was normalized to beta-actin.
Cell culture
Human colon adenocarcinoma cell lines (HT-29 and HCT116) and normal colon epithelial cell line (NCM460) were obtained from the Chinese Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultivated in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS, Gibco Waltham, MA), 100 U/mL penicillin and 100 μg/mL streptomycin (both from Sigma-Aldrich, St Louis, MO) at a humidified temperature of 37 °C with 5% CO2.
RNA transfection
Plasmids encoding specific RNAs interfering with LINC00491 (si-LINC00491) and corresponding RNAs interfering with scrambled (si-scrambled) vectors were synthesized by the RiboBio Corporation (Suzhou, China). miR-145 mimics and mimic controls were synthesized by RiboBio Corporation (Suzhou, China). siRNA plasmids were transfected into cells using Lipofectamine2000 (Invitrogen, Carlsbad, CA) transfection reagent according to manufacturers’ instructions. Six hours post-transfection, the medium was replaced with DMEM containing 10% FBS.
RNA extraction and real-time PCR analysis
Total RNAs were extracted using RNAiso Plus (TaKaRa, Tokyo, Japan). Reverse transcription kits (RR036A; TaKaRa) was used to transcribe total RNA and produce cDNA. Real-time PCR was performed with SYBR Premix Ex Taq II (TaKaRa) and NanoDrop 2000c (Thermo Scientific, Waltham, MA, USA). Gene expression was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6. Relative RNA expression was measured with ABI Prism 7500 Software v2.0.6 (Thermo Fisher Scientific, Waltham, MA) and calculated based on the 2−ΔΔC t method.
Western blot analysis
Proteins were isolated from tissues by lysing in radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich). Equal amounts of protein (50 μg) were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membranes (Thermo Fisher Scientific, Billerica, MA). Membranes were blocked in 5% bovine serum albumin (BSA) overnight and probed with primary antibodies against SERPINE1 and GAPDH (Cell Signaling Technology) as an internal control. Protein bands were visualized by enhanced chemiluminescence (ECL) (Applygen Technologies, Beijing, China).
Cell proliferation assays
Cells (100 μL/well) were seeded into 96-well plates at 37 °C, 5% CO2. CCK-8 solution (10 μL) was added to each well and plates were incubated for 24, 48, 72, and 96 hrs. Absorbances at 450 nm were measured on a microplate reader.
Colony formation assays
HT-29 and HCT116 cells were trypsinized to make a single cell suspension, 500 cells were inoculated into each well and the medium was changed every three days. Culture was stopped after 3 weeks, the cells were washed with phosphate-buffered saline (PBS), fixed with methanol and stained with crystal violet (Sinopharm Chemical Reagent, Beijing, China). The colonies were counted under a fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Wound healing assays
A horizontal line was evenly drawn across 6-well plates (Corning) every 0.5~1.0 cm and 5×105 cells were seeded into the wells. A cell scratch perpendicular to the horizontal line was made using a 200 μL pipette tip and cells were incubated at 37 °C, 5% CO2. Cells were imaged at 0, 24, and 48 hrs.
Transwell invasion assays
Cells (5×104 cells/200 μl of serum-free cell suspension) was added to Matrigel Invasion Chambers (size 8 μm; BD Biosciences, Franklin Lakes, NJ, USA), and 600 μL of medium containing 10% FBS was added to the lower chambers of 24-well plates (Corning). Cells were cultivated for 24 hrs, washed in PBS, fixed in methanol, and stained with crystal violet. Five cell fields were randomly counted on a fluorescent microscope (Olympus, Tokyo, Japan).
Dual luciferase reporter assays
To verify the binding sites of LINC00491-miR-145-SERPINE1, the psi-CHECK-2 vector (Promega, Madison, WI, USA) was cloned using XhoI and NotI sites. Mutant plasmids were generated by QuickChange® Site-Directed Mutagenesis (Stratagene, Agilent Technologies, Wilmington, DE, USA). Appropriate plasmid and miR-145 mimics or mimic controls were co-transfected into HEK293T cells (1.0×105), and luciferase activity was evaluated 48 hrs post-transfection using the Dual-luciferase Reporter Assay System (Promega). Values were normalized to Renilla luciferase activity.
Statistical analysis
Statistical analyses were performed using SPSS 20.0 software (IBM, Armonk, NY). Data are presented as the mean ± SD. Each experiment was performed in triplicate. Differences between the groups were determined using a two-tailed Student t-test. A one-way analysis of variance (ANOVA) was used for multiple comparisons. Kaplan-Meier methods and log rank tests were used to analyze the relationship between LINC00491 expression and overall survival rates of colon adenocarcinoma patients. The correlation of expression were analyzed through Spearman correlation analysis. p<0.05 was deemed significant.
Results
DEmRNAs, DEmiRNAs, and DElncRNAs in colon adenocarcinoma samples
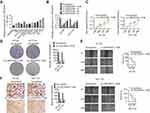
The RNA expression profiles of patients with colon adenocarcinoma and corresponding their corresponding clinical information were downloaded from the TCGA database using the Data Transfer Tool. EdgeR was used to identify significant differences in DEmRNAs, DEmiRNAs, and DElncRNAs between colon adenocarcinoma and normal samples (adjusted p-value<0.01 and |logFC|>2 as the thresholds). We identified a total of 2081 DEmRNAs, 245 DEmiRNAs, and 1176 DElncRNAs. More specifically, 1170 (56.2%) were up-regulated and 911 (43.8%) were down-regulated DEmRNAs (Table S1), 174 (71.0%) were up-regulated and 71 (29.0%) were down-regulated DEmiRNAs (Table S2), and 912 (77.6%) were up-regulated and 264 (22.4%) were down-regulated DElncRNAs (Table S3). Heat maps are shown in Figure S1, and volcano plot demonstrating the clustering of DElncRNAs, DEmiRNAs, and DEmRNAs is shown in Figure 1A–C.
Construction of the ceRNA network
To reveal how lncRNAs mediate transcription in colon adenocarcinoma through their influence on mRNA and miRNA binding, a ceRNA network based on the list of DElncRNAs, DEmiRNAs, and DEmRNAs were constructed and visualized using Cytoscape software. As shown in Figure S2, the lncRNA-miRNA-mRNA network was comprised of 135 lncRNA, 28 miRNA and 52 mRNA nodes (Table S4–5). The majority of DEmRNAs were tumor-related genes including SERPINE1, NPTX1, HILPDA, PRSS22, PMAIP1.12–14 Interestingly, all formed a ceRNA network with LINC00491 (Figure 1D).
Predicated functions of mRNAs within the lncRNA–miRNA–mRNA network
Functional analysis of lncRNAs and mRNAs of the ceRNA network was performed using GO terms and KEGG pathways. We selected the top 13 most significantly enriched GO terms (Figure 1E). The top 12 KEGG pathways are shown in Figure 1F. GO analysis was performed for the assessment of the SERPINE1 signaling axis (Table 1). Amongst the pathways identified, protease binding, fibrinolysis, and platelet degranulation have been shown to be involved in cancer pathogenesis.15–19
 |
Table 1 SERPINE1 enriched GO terms |
LINC00491 promotes colon adenocarcinoma cell proliferation, migration and invasion
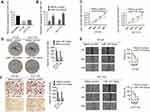
Real-time PCR showed that LINC00491 was overexpressed in colon adenocarcinoma cell lines compared to normal colon cells (NCM460) (Figure 2A). HT-29 and HCT116 cells were chosen to study the function of LINC00491 in cell culture. When si-LINC00491-1#, si-LINC00491-2#, si-LINC00491-3#, si-LINC00491-123# or scrambled were transfected into HT-29 and HT116 cells, LINC00491 expression was remarkably reduced in si-LINC00491-123# compared to scrambled controls (Figure 2B). When the proliferation of transfected HT-29 and HCT116 cells was evaluated by CCK-8 and plate clone formation assays, si-LINC00491-123# decreased cell proliferation (Figure 2C and D). We reasoned that LINC00491 may control the migration and invasion of colon adenocarcinoma cells. To investigate this, wound healing and transwell invasion assays were performed in HT-29 and HCT116 cells transfected with si-LINC00491-123# or scrambled controls. The results showed that cell migration and invasion ability were reduced in si-LINC00491-123# transfected cells (Figure 2E and F). These data suggest that LINC00491 is an oncogene that promotes colon adenocarcinoma cell proliferation, migration and invasion.
miR-145 functions as a suppressor gene in colon adenocarcinoma
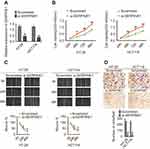
We used RT-PCR to detect miR-145 levels in HT-29, HCT116, and NCM460 cells. In contrast to LINC00491, miR-145 was significantly suppressed in HT-29 and HCT116 cells (Figure 3A), highlighting its inhibition in colon adenocarcinoma. The levels of miR-145 increased in miR-145 mimic transfected HT-29 and HCT116 cells (Figure 3B) whilst the proliferation of both cell lines decreased (Figure 3C). Increasing the expression of miR-145 also reduced plate clone formation compared to mimic controls (Figure 3D). Cell migration was also inhibited in miR-145 mimic transfected HT-29 and HCT116 cells compared to mimic controls (Figure 3E). Transwell invasion assays demonstrated that invasiveness was significantly reduced following miR-145 mimic transfection (Figure 3F). Taken together, these data indicate anti-cancer effects of miR-145 in colonic adenocarcinoma cells.
LINC00491 ceRNA activity regulates SERPINE1 expression
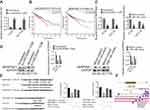
HT-29 and HCT116 cells were transfected with SERPINE1 siRNA and silencing was confirmed by qRT-PCR (Figure 4A). CCK8 assays showed that SERPINE1 silencing significantly reduced colon adenocarcinoma cells growth and viability (Figure 4B). Moreover, SERPINE1 silencing could inhibit colon adenocarcinoma cell migration (Figure 4C). In addition, HT-29 and HCT116 cells transfected with SERPINE1 siRNA decreased the invasion ability of colon adenocarcinoma cells (Figure 4D).
To study the role of SERPINE1 in colon adenocarcinoma, cells were transfected with LINC00491-miR-145-SERPINE1 and RT-PCR assays was used to determine the expression of miR-145. miR-145 expression significantly increased in si-LINC00491-123# transfected cells compared with scrambled controls (Figure 5A). Through Kaplan-Meier curve analysis, the expression of LINC00491 and SERPINE1 negatively correlated with the overall survival rates (Figure 5B). LINC00491 was found to regulate SERPINE1 levels, since transfection with si-LINC00491-123# decreased SERPINE1 transcript and protein levels in HT-29 and HCT116 cells. RT-PCR and Western blot analysis showed that SERPINE1 expression was reduced in HT-29 and HCT116 cells transfected with miR-145 mimics compared to controls (Figure 5C and D). In addition, we constructed a series of luciferase reporter genes containing WT SERPINE1 3’UTR, mutant SERPINE1 3’UTR, wild LINC00491 or mutant LINC0049 that were co-transfected into cells with LINC00491-miR-145-SERPINE 3’UTR or miR-145 mimics. We found that the miR-145-mediated inhibition of luciferase activity was abolished by mutations in LINC00491, and that WT-SERPINE1 3’UTR driven luciferase expression decreased following co-transfection of the miR-145 mimic (Figure 5E). These results indicate that LINC00491 regulates SERPINE1 through its competitive binding to miR-145.
Discussion
Colon adenocarcinoma is a common malignant tumor and one of the leading causes of cancer-related death worldwide.20 The lack of specific diagnostic and prognostic biomarkers results in low survival rates in colon adenocarcinoma patients.21 It is therefore important to discover novel and effective colon adenocarcinoma treatment targets.
Increasing evidence suggests that lncRNA expression profiles can be used as molecular markers for tumor diagnosis and prognosis, including breast cancer,22 gastric cancer,23 hepatocellular carcinoma,24 and colon adenocarcinoma.25 In this study, RNA sequencing data and the clinical characteristics of colon adenocarcinoma were obtained from the Cancer Genome Atlas database. DElncRNAs, DEmRNAs, and DEmiRNAs were identified between colon adenocarcinoma and normal colon tissue samples. Subsequently, the lncRNA-miRNA-mRNA ceRNA network of colon adenocarcinoma was established. We found that LINC00491 acts as a key regulator of human colon adenocarcinoma and revealed new roles of the LINC00491-miR-145-SERPINE1 axis. Interestingly, Liu et al showed that LINC00491 is associated with overall survival rates in breast cancer patients.26 Wang et al used the TCGA database and reported the upregulation of LINC00491 and the downregulation of miR-145. A link between LINC00491, poor prognosis and SERPINE1 expression was also reported, consistent with our findings.27 MiRNAs are abnormally expressed in tumors and represent novel therapeutic approaches for tumor therapy. miR-145 is down-regulated in several cancers, including colon adenocarcinoma28 and thyroid cancer.29 SERPINE1 is significantly upregulated in gastric tissue and is associated with a poor outcome.30 To determine whether LINC00491 is involved in the tumorigenesis of colon adenocarcinoma, real-time PCR was used to detect the expression of LINC00491 in colon adenocarcinoma cell lines. Our results demonstrate that LINC00491 is highly expressed in colon adenocarcinoma cell lines. Functional assays showed that si-LINC00491 and SERPINE1 silencing decrease cell proliferation, migration, and the invasion of HT29 and HCT116 cells, which can be recapitulated using mimic-miR-145. The ceRNA network are important regulators and lncRNAs function as ceRNAs through their ability to regulate miRNAs in human cancers.31–33 We therefore investigated whether LINC00491 acts as a ceRNA in colon adenocarcinoma through RT-PCR and Western blot analysis, and found that miR-145 was up-regulated, but SERPINE1 was down-regulated in colon adenocarcinoma cells after treatment with si-LINC00491. We performed dual luciferase reporter assay to confirm that LINC00491 functions as a ceRNA and competitively binds to miR-145, up-regulating SERPINE1. Taken together, these results indicate that lncRNA LINC00491 promotes colon adenocarcinoma cell proliferation, migration and invasion by competitively binding to miR-145, up-regulating SERPINE1, and accelerating colon adenocarcinoma metastasis (Figure 5F).
In summary, this study demonstrates that LINC00491 functions as a ceRNA to promote SERPINE1 expression through its ability to sponge miR-145. LINC00491 may therefore serve as a therapeutic target and prognostic biomarker in colon adenocarcinoma.
Acknowledgment
This work was supported by grants from the Postgraduate Innovation Research Project of Mudanjiang Medical University (No.2019YJSCX-02MY).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Byrne M, Saif MW. Selecting treatment options in refractory metastatic colorectal cancer. Onco Targets Ther. 2019;12:2271–2278. doi:10.2147/OTT.S194605
2. Fu X, Huang Y, Fan X, et al. Demographic trends and KRAS/BRAFV600E mutations in colorectal cancer patients of South China: a single-site report. Int J Cancer. 2018. [Epub ahead of print]. doi:10.1002/ijc.31973
3. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality wordwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
4. Raveendra BL, Swarnkar S, Avchalumov Y, et al. Long noncoding RNA GM12371 acts as a transcriptional regulator of synapse function. Proc Natl Acad Sci USA. 2018;115(43):E10197–E10205. doi:10.1073/pnas.1722587115
5. Li Y, Zhou L, Lu C, et al. Long non-coding RNA FAL1 functions as a ceRNA to antagonize the effect of miR-637 on the down-regulation of AKT1 in Hirschsprung’s disease. Cell Prolif. 2018;51:e12489. doi:10.1111/cpr.12489
6. Dong Z, Zhang A, Liu S, et al. Aberrant methylation-mediated silencing of lncRNA MEG3 functions as a ceRNA in Esophageal cancer. Mol Cancer Res. 2017;15:800–810. doi:10.1158/1541-7786.MCR-16-0385
7. LIU T, CHI H, CHEN J, et al. Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of MiR-145 and LncRNA-ROR. Gene. 2017;631:29–38. doi:10.1016/j.gene.2017.08.008
8. Ye Y, Gu B, Wang Y, et al. E2F1-mediated MNX1-AS1-miR-218-5p-SEC61A1 feedback loop contributes to the progression of colon adenocarcinoma. J Cell Biochem. 2018. [Epub ahead of print]. doi:10.1002/jcb.27902
9. Xu Y, Yao Y, Qin W, et al. Long non-coding RNA CCAT2 promotes cholangiocarcinoma cells migration and invasion by induction of epithelial-to-mesenchymal transition. Biomed Pharmacother. 2018. [Epub ahead of print]. doi:10.1016/j.biopha.2018.01.061
10. Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi:10.1186/s12943-017-0680-1
11. Yang X, Song JH, Cheng Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi:10.1136/gutjnl-2013-305266
12. Liang R, Xiao G, Wang M, et al. SNHG6 functions as a competing endogenous RNA to regulate E2F7 expression by sponging miR-26a-5p in lung adenocarcinoma. Biomed Pharmacother. 2018;107:1434–1446. doi:10.1016/j.biopha.2018.08.099
13. Bian Y, Guo J, Qiao L, et al. miR-3189-3p mimics enhance the effects of S100A4 siRNA on the inhibition of proliferation and migration of gastric cancer cells by targeting CFL2. Int J Mol Sci. 2018;19:
14. Feng A, Yuan X, Li X. MicroRNA-345 inhibits metastasis and epithelial-mesenchymal transition of gastric cancer by targeting FOXQ1. Oncol Rep. 2017;38:2752–2760. doi:10.3892/or.2017.6001
15. Deng Y, He R, Zhang R, et al. The expression of HOXA13 in lung adenocarcinoma and its clinical significance: a study based on The Cancer Genome Atlas, Oncomine and reverse transcription-quantitative polymerase chain reaction. Oncol Lett. 2018;15:8556–8572. doi:10.3892/ol.2018.8381
16. Campos A, Salomon C, Bustos R, et al. Caveolin-1-containing extracellular vesicles transport adhesion proteins and promote malignancy in breast cancer cell lines. Nanomedicine (Lond). 2018;13:2597–2609. doi:10.2217/nnm-2018-0094
17. Banda NK, Desai D, Scheinman RI, et al. Targeting of liver mannan-binding lectin-associated serine protease-3 with RNA interference ameliorates disease in a mouse model of rheumatoid arthritis. Immunohorizons. 2018;2:274–295. doi:10.4049/immunohorizons.1800053
18. Farag M, Spinthakis N, Gue YX, et al. Impaired endogenous fibrinolysis in ST-segment elevation myocardial infarction patients undergoing primary percutaneous coronary intervention is a predictor of recurrent cardiovascular events: the RISK PPCI study. Eur Heart J. 2018. [Epub ahead of print]. doi:10.1093/eurheartj/ehy656
19. Serhan K, Gartung A, Panigrahy D. Drawing a link between the thromboxane A2 pathway and the role of platelets and tumor cells in ovarian cancer. Prostaglandins Other Lipid Mediat. 2018;137:40–45. doi:10.1016/j.prostaglandins.2018.06.001
20. LIN J, SHI Z, YU Z, et al. LncRNA HIF1A-AS2 positively affects the progression and EMT formation of colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed Pharmacother. 2018;98:433–439. doi:10.1016/j.biopha.2017.12.058
21. Yan L, Yao J, Qiu J. miRNA-495 suppresses proliferation and migration of colorectal cancer cells by targeting FAM83D. Biomed Pharmacother. 2017;96:974–981. doi:10.1016/j.biopha.2017.11.138
22. Yang F, Shen Y, Zhang W, et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ. 2018. [Epub ahead of print]. doi:10.1038/s41418-018-0123-6
23. Peng W, Si S, Zhang Q, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi:10.1186/s13046-015-0197-7
24. Wang Y, Sun L, Wang L, et al. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/β-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis. 2018;9:851. doi:10.1038/s41419-018-1111-y
25. Chen DL, Lu YX, Zhang JX, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849. doi:10.7150/thno.20942
26. Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med. 2018;16(1):264. doi:10.1186/s12967-018-1640-2
27. Wang WJ, Li HT, Yu JP. A competing endogenous RNA network reveals novel potential lncRNA, miRNA, and mRNA biomarkers in the prognosis of human colon adenocarcinoma. J Surg Res. 2019;235:22–33.
28. Ibrahim AF, Weirauch U, Thomas M, et al. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71(15):5214–5224. doi:10.1158/0008-5472.CAN-10-4645
29. Li X, Tian Y, Hu Y, et al. CircNUP214 sponges miR-145 to promote the expression of ZEB2 in thyroid cancer cells. Biochem Biophys Res Commun. 2018;
30. Liao P, Li W, Liu R, et al. Genome-scale analysis identifies SERPINE1 and SPARC as diagnostic and prognostic biomarkers in gastric cancer. Onco Targets Ther. 2018;11:6969–6980. doi:10.2147/OTT.S173934
31. Wang Q, Cai J, Fang C, et al. Mesenchymal glioblastoma constitutes a major ceRNA signature in the TGF-β pathway. Theranostics. 2018;8:4733–4749. doi:10.7150/thno.26550
32. Liu HT, Liu S, Liu L, et al. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell-cycle progression in gastric cancer. Cancer Res. 2018;78:5877–5890. doi:10.1158/0008-5472.CAN-18-1011
33. Ma R, Zhai X, Zhu X, et al. LINC01585 functions as a regulator of gene expression by the CAMP/CREB signaling pathway in breast cancer. Gene. 2018;684:139–148. doi:10.1016/j.gene.2018.10.063
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.