Back to Journals » International Journal of Nanomedicine » Volume 15
Lethal Mechanisms of Nostoc-Synthesized Silver Nanoparticles Against Different Pathogenic Bacteria
Authors Hamida RS , Ali MA, Goda DA, Al-Zaban MI
Received 28 October 2020
Accepted for publication 5 December 2020
Published 29 December 2020 Volume 2020:15 Pages 10499—10517
DOI https://doi.org/10.2147/IJN.S289243
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Thomas Webster
Reham Samir Hamida,1 Mohamed Abdelaal Ali,2 Doaa A Goda,3 Mayasar Ibrahim Al-Zaban4
1Molecular Biology Unit, Department of Zoology, Faculty of Science, Alexandria University, Alexandria, Egypt; 2Biotechnology Unit, Department of Plant Production, College of Food and Agriculture Science, King Saud University, Riyadh, Saudi Arabia; 3Bioprocess Development Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), Alexandria, Egypt; 4Department of Biology, College of Science Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
Correspondence: Mohamed Abdelaal Ali Email [email protected]
Mayasar Ibrahim Al-Zaban Email [email protected]
Background: Increasing antibiotic resistance and the emergence of multidrug-resistant (MDR) pathogens have led to the need to develop new therapeutic agents to tackle microbial infections. Nano-antibiotics are a novel generation of nanomaterials with significant antimicrobial activities that target bacterial defense systems including biofilm formation, membrane permeability, and virulence activity.
Purpose: In addition to AgNO3, the current study aimed to explore for first time the antibacterial potential of silver nanoparticles synthesized by Nostoc sp. Bahar_M (N-SNPs) and their killing mechanisms against Streptococcus mutans, methicillin-resistant Staphylococcus aureus, Escherichia coli, and Salmonella typhimurium.
Methods: Potential mechanisms of action of both silver species against bacteria were systematically explored using agar well diffusion, enzyme (lactate dehydrogenase (LDH) and ATPase) and antioxidant (glutathione peroxidase and catalase) assays, and morphological examinations. qRT-PCR and SDS-PAGE were employed to investigate the effect of both treatments on mfD, flu, and hly gene expression and protein patterns, respectively.
Results: N-SNPs exhibited greater biocidal activity than AgNO3 against the four tested bacteria. E. coli treated with N-SNPs showed significant surges in LDH levels, imbalances in other antioxidant and enzyme activities, and marked morphological changes, including cell membrane disruption and cytoplasmic dissolution. N-SNPs caused more significant upregulation of mfD expression and downregulation of both flu and hly expression and increased protein denaturation compared with AgNO3.
Conclusion: N-SNPs exhibited significant inhibitory potential against E. coli by direct interfering with bacterial cellular structures and/or enhancing oxidative stress, indicating their potential for use as an alternative antimicrobial agent. However, the potential of N-SNPs to be usable and biocompatible antibacterial drug will evaluate by their toxicity against normal cells.
Keywords: biological synthesis, nanoparticles, pathogenic bacteria, Nostoc sp. Bahar_M, silver, antibacterial activity
Introduction
Emerging infectious diseases caused by pathogenic microbes is a global health crisis.1 The surge in antibiotic-resistant bacterial infections poses health risks to humans and animals, and impacts the global economy.2 Pathogenic bacteria and multidrug-resistant (MDR) bacteria, or superbugs, have adapted and modified their defense strategies against antibiotics over time.3 For instance, resistant bacteria can mitigate outer membrane permeability to prevent antibiotics entering cells, and can also form biofilm as a protective barrier against antimicrobial agents.4–6 Moreover, bacteria utilize a variety of different virulence factors to invade hosts and cause infections, including adherence and invasion factors, capsules, and endo- and exo-toxins.7,8 These defense strategies can inhibit the efficacy of antimicrobial agents in tackling MDR microbes and increase the need for new alternate strategies to alleviate the spread and progression of MDR pathogens.9 One approach to mitigate MDR microbes is to utilize nano-antibiotics.10,11
Nanotechnology facilities many options to develop and create antimicrobial drugs in trial to tackle the MDR microbes’ phenomena.12–14 Many nanodrugs have been approved by the Food and Drug Administration for use as burns therapy (e.g., Acticoat), as antifungal and anti-parasitic agents (e.g., Abelcet® and AmBIsome).15,16 The potentiality of nanoparticles (NPs) to be promising antimicrobial agents is returning to their physical and chemical properties.17 The smaller size of NPs enables them to easily penetrate the cells such as bacteria causing cellular damage.18 Additionally, larger surface area of NPs allows them to be load with antibiotics and antimicrobial agents increasing their efficiency;19 their charges play important role in nano/cell interfere;20 their stability,19 biocompatibility and bioavailability enabling them to be used in different medical applications.21,22
There are several methods to synthesize NPs including conventional physical and chemical approaches and through ecofriendly routes such as biological synthesis.23 Unlike chemical methods, biological synthesis of NPs uses natural sources such as bacteria, plant, cyanobacteria, etc. to fabricate metal precursors into their nanoforms.17,21,24–26 Thus, green synthesis approach does not produce or use toxic materials, making it safer for living organisms and the environment.27,28 Similarly, biological synthesis of NPs has become more attractive than physical synthesis methods because biological synthesis can easily be performed under normal laboratory conditions without expensive instruments, intensive power, or significant costs.17,29 Recently, many investigations used cyanobacteria as a bio-factory for synthesizing NPs showing the higher reducing potentiality of these creatures to fabricate the precursors material into their nanoforms.27,30 Hamouda et al reported that Ag-NPs synthesized by Oscillatoria limnetica revealing potent anticancer and antibacterial potentiality.31
In the medical field, metallic nanoparticles such as silver NPs (Ag-NPs),32 and gold NPs,33 and metal oxide NPs such as zinc oxide NPs,34 and titanium oxide NPs play critical roles as therapeutic agents against cancers and infectious diseases, including viral and microbial infections.35–38 Among the metallic NPs, Ag-NPs have been extensively studied due to their wide-ranging biological activities, including as potent antibacterial,20,39 anti-fungal,40 anti-larval,41 anti-viral,42 anti-cancer,43 and anti-inflammation agents.44 Junejo et al reported that tobramycin stabilized Ag-NPs exhibiting potent antibacterial activity against Escherichia coli and Staphylococcus aureus comparing to the standard antibiotics.45 Furthermore, Ag-NPs have unique physicochemical characteristics, including a very small size and large surface area, allowing them to easily penetrate living cells and making them suitable for use in drug delivery and synergetic and antagonistic technology.9,46–48 Ruden et al reported that the combination between Ag-NPs and polymyxin B exhibiting the most promising antibiotic synergy against Gram-negative bacteria.49
Many investigations have focused on the synthesis of NPs, but the exact mechanism of action of NPs against living cells including microbes, healthy human cells, and cancer cells has yet to be elucidated.43,50 There are two general theories regarding the mechanism of the lethal effects of NPs against bacterial cells. The first theory is that NPs directly interfere with cellular structures including cell walls, cell membranes, and the cytoplasmic matrix through electrostatic interactions, causing membrane disruption, folding, and pore formation. This results in increased membrane permeability, loss of membrane integrity, and bacterial cell death, with denaturation, damage, and dysfunction of cellular biomolecules (DNA, protein, and enzymes).20,51 The second theory is that NPs enhance the formation of reactive oxygen species (ROS) leading to a surge in oxidative stress, which causes damage to biomolecules and cell structures, and ultimately, bacterial cell death.20,35,52,53
The current study aimed to investigate for first time the antibacterial activity and mechanisms of silver nanoparticles (synthesized by novel cyanobacteria Nostoc sp. Bahar_M) enhanced cytotoxicity against different pathogenic bacteria including Gram-positive (Streptococcus mutans and methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates) and Gram-negative (Escherichia coli ATCC 25,922 and Salmonella typhimurium ATCC 14,028) pathogenic bacteria, comparing to that of silver nitrate (AgNO3; a precursor material for Ag-NP synthesis). Depend on the results of the current study, we speculated that N-SNPs are more robust and potent antibacterial candidate against the pathogenic bacteria.
Materials and Methods
Reagents
AgNO3, bacterial culture materials, and catalase (CAT) and glutathione peroxidase (GPx) enzymes were purchased from Sigma-Aldrich (St. Louis, MO, USA). Other reagents were purchased from the following companies: LDH Assay Kit (colorimetric) from Abcam (Cambridge, UK); PiBind resin from Expedeon (San Diego, USA); TRIzol reagent from Life Technologies (California, USA); Maxima SYBR Green/Fluorescein qPCR Master Mix and QuantiTects Reverse Transcription Kit from Qiagen (Germantown, USA); and TriFast from Peqlab VWR (Pennsylvania, USA).
Preparation of N-SNP and AgNO3 Suspensions
Ag-NPs were previously synthesized extracellularly by Nostoc sp. Bahar_M and characterized by UV-spectrophotometry, X-ray diffraction, Fourier transform infrared spectroscopy (FTIR), and scanning and transmission electron microscopy (TEM).27 Briefly, Nostoc sp. biomass was collected by centrifugation at 4000 rpm. Pellets were washed with distilled water several times, freeze-dried using a lyophilizer, and ground into a fine powder using a mortar and pestle. Subsequently, 20 mg dried powder was mixed with 20 mL distilled water and incubated at 30°C for 24 h. After cooling, the mixture was filtered using Whatman filter paper no.1 (Camlab, Cambridge, UK) and 10 mL filtrate was added to 90 mL AgNO3 solution and stored at room temperature for 24 h in the dark. The mixture was then centrifuged at 10,000 rpm for 10 min. The resulting pellets were washed with distilled water at least three times and spread on sterilized glass plates to dry at 40°C for 24 h. Stock N-SNPs and AgNO3 solutions were prepared by dissolving 1 mg of each silver reagent in 1 mL of distilled water.
Bacterial Strains and Culture Conditions
Four different pathogenic bacteria, including clinical isolates of Streptococcus mutans and methicillin-resistant Staphylococcus aureus (MRSA), and E. coli ATCC 25,922 and Salmonella typhimurium ATCC 14,028 were obtained from Almery University Hospital (Alexandria, Egypt). Fresh bacterial cultures (104 CFU/mL), at a concentration of 0.5 on the McFarland scale, were grown in Luria–Bertani (LB) broth for 24 h at 37°C. Each strain (50 µL) was gently spread on freshly prepared LB agar plates. Four 8-mm wells were created using a cork borer in the agar of each plate for further antibacterial activity experiments.
Antibacterial Potentials of N-SNPs and AgNO3
The inhibitory effect of N-SNPs and AgNO3 on Streptococcus mutans, MRSA, E. coli, and Salmonella typhimurium was assessed using the agar well diffusion method.54 One hundred microliters of each N-SNPs, AgNO3 (1.5 mg/mL), distilled water (as a negative control), and ampicillin (1.5 mg/mL) (as a positive control) were added to the wells of agar plates spread with bacterial cultures. Plates were incubated for 24 h at 37°C, then the diameter of each inhibition zone was recorded in mm using a standard metric ruler.
Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
Minimum inhibitory and minimum bactericidal concentrations (MIC and MBC, respectively) of both N-SNPs and AgNO3 were evaluated using the serial dilution method. Bacterial suspensions (100 µL/well of 104 CFU/mL) were seeded in 96-well plates with different N-SNPs and AgNO3 concentrations (2.4, 2.1, 1.8, 1.5, 1.2, 0.9, 0.6, and 0.3 mg/mL) and incubated at 37°C for 24 h. Bacterial turbidity was estimated by naked eye comparison to a 0.5 McFarland standard medium. To verify the MIC and MBC data, 10 µL N-SNPs or AgNO3 solution at the determined MIC and MBC were screened against the tested bacteria using an agar well diffusion assay.
Lactate Dehydrogenase Assay for Membrane Integrity
The effect of 1.5 mg/mL N-SNPs or AgNO3 on bacterial membrane integrity was assessed by measuring lactate dehydrogenase (LDH) as previously described.55 Bacterial cells (104 CFU/mL) treated with 1.5 mg/mL N-SNPs or AgNO3 for 24 h at 37°C, and corresponding controls, were collected by centrifugation at 5000 rpm for 10 min at 4°C. The resulting pellets were washed twice with phosphate-buffered saline (PBS) and mixed with LDH reaction solution under gentle shaking for 30 min at room temperature according to manufacturer’s instructions. The sample optical density (OD) was then measured at 490 nm.
ATPase Activity
The effect of 1.5 mg/mL N-SNPs and AgNO3 on metabolism was estimated by measuring bacterial ATPase levels before and after exposure to both treatments for 24 h, as previously described.56 Briefly, cells were centrifuged at 5000 rpm for 10 min at 4°C. The pellets were washed with PBS and lysed by sonication (IKAT10 basic sonicator, Cole-Parmer, Illinois, USA). PiBind resin was used to remove any free inorganic phosphate (Pi) that would interfere with the ATPase assay. The amount of Pi liberated was evaluated with a UV 2505 spectrophotometer (Thomas Scientific, New Jersey, USA) at A650. Calibration was based on a standard Pi concentration range, and data were obtained from a minimum of three independent tests.
Measurement of Antioxidative Markers
To assess the impact of N-SNPs or AgNO3 on bacterial antioxidant enzymes, including glutathione peroxidase (GPx) and catalase (CAT), bacterial cells treated with both silver species (1.5 mg/mL) for 24 h at 37°C, and appropriate controls, were centrifuged at 10,000 rpm for 5 min at 4°C. Pellets were washed with PBS and lysed using a sonicator (IKAT10 basic sonicator, Cole-Parmer, Illinois, USA). GPx and CAT levels were determined using reagents from various kits according to the corresponding instructions.57
Ultrastructural Examination
E. coli cultures exposed to 1.5 mg/mL of N-SNPs or AgNO3 for 24 h at 37°C, and appropriate controls, were harvested by centrifugation at 3500 rpm for 10 min. The resulting pellets were washed several times with PBS and recentrifuged. Subsequently, pellets were fixed with ice-cold 4F1G (formalin-glutaraldehyde mixture (pH 7.2)) for 2 h, then post-fixed with 1% osmium tetroxide (OsO4) for 2 h at 4°C. The specimens were dehydrated with graded ethanol (25%, 50%, 75%, 95%, and 100%), infiltrated with propylene oxide, and embedded in an Araldite Epon mixture. Ultrathin sample sections (70 nm) were created using a glass knife on an LKB Ultramicrotome. The sections were double stained using 2% uranyl acetate and lead citrate, and loaded on 200-mesh copper grids for morphological examination under a JEOL 100 CX electron microscope (JEOL, Tokyo, Japan) operating at 80 kV.58
Size Estimation of NPs and Bacterial Cells
The size of NP inside and outside of bacterial cells and bacterial cell size before and after exposure to N-SNPs and AgNO3 were measured by estimating the NPs and bacterial cell diameter based on TEM micrographs using ImageJ software (National Institutes of Health, Bethesda, MD, USA); at least 10 bacterial cells were used for measurements.
RNA Extraction and qRT-PCR
Expression of the genes mfD (transcription-repair-coupling factor), hly (α-hemolysin), and flu (Ag43 phase-variable biofilm formation autotransporter CP4-44 prophage) in E. coli were estimated using qRT-PCR before and after treatment with 1.5 mg/mL N-SNPs or AgNO3 for 24 h at 37°C (Table 1). Total RNA was extracted from samples using TRIzol reagent. The amount and purity of the RNA was determined by measuring absorbance at 260 nm and by the 260/280 nm ratio, respectively. mfD, hly, and flu mRNA was assessed using Maxima SYBR Green/Fluorescein qPCR Master Mix and a Rotor-Gene Q instrument. Total RNA was reverse-transcribed using QuantiTects Reverse Transcription Kit with a random primer hexamer in a two-step RT-PCR. Any genomic DNA (gDNA) contamination was eliminated using gDNA Wipeout Buffer. The resultant cDNA (30 ng) was used as a template for amplification using specific primer pairs (Table 1) at a final concentration of 300 nM. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene. The Rotor-Gene Q automatically assembled the data and analyzed the threshold cycle (Ct) value, which was normalized to the average Ct value of the housekeeping gene (ΔCt), and relative expression of each assessed gene was calculated as 2–ΔCt.54
 |
Table 1 qRT-PCR Primers |
SDS-PAGE
The influence of N-SNPs and AgNO3 on bacterial proteins was investigated by collecting total proteins from E. coli cells that were untreated or treated with 1.5 mg/mL of N-SNPs or AgNO3 for 24 h, and from appropriate controls. Proteins were purified using TriFast and fractionated using an OmniPAGE Mini vertical electrophoresis unit with a Power Pro 5 power supply (Cleaver Scientific, Warwickshire, UK) on a SERVAGel™ TG PRiME™ 10% (SERVA, Heidelberg, Germany). The gel was stained with 0.1% Coomassie blue R250 for 2 h and de-stained with a 1:3:6 solution of glacial acetic acid:methanol:water. A gel documentation system (Geldoc-it, UVP, UK) and TotalLab analysis software version 1.0.1 (Newcastle-Upon-Tyne, UK) were used for data analysis.54
Statistical Analysis
All assays were performed independently at least three times and data are presented as mean ± standard error of mean (SEM). Significant differences between the study data (treated and untreated samples) was statistically determined using one-way analysis of variance (ANOVA) and Prism 8.3 software (GraphPad Software Inc., La Jolla, CA, USA). Significance of the data is presented at P < 0.0001, P = 0.0002, and P < 0.001.
Results
N-SNPs
Our previous published data showed that Nostoc Bahar _M successfully fabricated silver nitrate into N-SNPs. UV-spectra of N-SNPs was at 403 nm, while XRD pattern revealed that N-SNPs have crystalline nature at 2θ of 38.2, 45.3, 67.44, and 75.25° and with crystal size of 3.8 nm. FTIR analysis of N-SNPs exhibited spectra peaks at 1119.01, 1397.07, 1632.35, 1777.14, 2114.44, 2946.98, 3460.32 and 842.59 cm−1 which corresponding to (C-O) secondary alcohol, (O-H) carboxylic acid, (N-H) stretching amine, (C=O) stretching anhydride or vinyl/phenyl ester, (N=C=S) stretching isothiocyanate, (O-H) carboxylic acid or (N-H) amine salt, (O-H) stretching alcohol, and (C=C) stretching alkene, respectively. These IR spectra indicated that the aromatic compounds and proteins of Nostoc sp. was the main biomolecules in the reduction and stabilization processes of NPs. TEM and SEM micrographs exhibited that N-SNPs having spherical shape with a nanosize range of 8.5 nm to 26.44 nm, and an average diameter of 14.9 ± 0.56 nm (Figure 1).27
Inhibitory Activity of Both Silver Species Against Pathogenic Bacteria
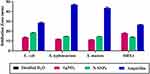
The bactericidal effect of N-SNPs, AgNO3, and ampicillin against Gram-positive and Gram-negative bacterial strains was investigated using the agar well diffusion method. All treatments demonstrated bactericidal activity against the tested bacteria, with ampicillin producing the largest inhibition zone (IZ) diameter, followed by N-SNPs then AgNO3 (Figure 2). The bacterial inhibitory activity of N-SNPs was greater than that of AgNO3 (Figure 3). Of the tested bacteria, E. coli and MRSA were most affected by N-SNPs, with IZ diameters of 18.6 ± 0.07 and 18 ± 0.09 mm, respectively (E. coli ˃ MRSA ˃ Salmonella typhimurium ˃ Streptococcus mutans, Table 2). In contrast, MRSA was the most sensitive to AgNO3 (IZ diameter: 17.5 ± 0.27 mm) followed by E. coli (IZ diameter: 13.7 ± 0.29 mm). Salmonella typhimurium and Streptococcus mutans had similar IZ diameters against N-SNPs (14.8 ± 0.06 and 14.7 ± 0.03 mm, respectively) and AgNO3 (11.9 ± 0.15 and 11.4 ± 0.12 mm, respectively). N-SNPs had lower MIC and MBC values than AgNO3 against the tested bacteria (Table 3). The lowest N-SNPs MIC (0.9 mg/mL) was against Salmonella typhimurium, E. coli and MRSA while the lowest MBC (1.2 mg/mL) was against both E. coli and Salmonella typhimurium. The highest MIC of N-SNPs was 1.2 mg/mL against Streptococcus mutans, while the highest MBC (1.5 mg/mL) was against both MRSA and Streptococcus mutans, respectively. In contrast, the lowest AgNO3 MIC was 1.5 mg/mL against E. coli, MRSA, and Streptococcus mutans, while the lowest MBC was 1.8 mg/mL against MRSA and Streptococcus mutans. Moreover, the highest AgNO3 MIC and MBC values were 2.1 and 2.4 mg/mL, respectively, against Salmonella typhimurium (Table 3).
 |
Table 2 Inhibitory Activity of N-SNPs, AgNO3, and Ampicillin Against Four Pathogenic Bacteria |
 |
Table 3 Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of N-SNPs and AgNO3 Against Four Pathogenic Bacteria |
Effect of N-SNPs and AgNO3 on Bacterial Membrane Integrity
The influence of N-SNPs and AgNO3 on bacterial membrane integrity was examined by measuring LDH in bacterial supernatants treated with the silver species. Bacteria exposed to 1.5 mg/mL N-SNPs and AgNO3 showed a significant increase in LDH compared to untreated bacterial cells, with N-SNPs treatment producing the most significant increase in LDH (Figure 4A). Of the four tested bacterial species, E. coli had the highest level of LDH after treatment with N-SNPs, while AgNO3 treatment produced the highest level of LDH in MRSA.
Metabolic Toxicity of N-SNPs and AgNO3
The metabolic toxicity of N-SNPs and AgNO3 on bacteria was assessed by measuring ATPase levels before and after treatment. The amount of ATPase significantly decreased in E. coli, Salmonella typhimurium, and MRSA after treatment with N-SNPs (Figure 4B), but ATPase levels increased in Streptococcus mutans after N-SNPs treatment. AgNO3 treatment caused a similar significant decrease in amounts of ATPase in the examined bacteria. Of the four bacterial species tested, the greatest reduction in ATPase was observed in E. coli treated with N-SNPs.
Enhancement of Oxidative Stress by N-SNPs and AgNO3
The ability of N-SNPs and AgNO3 to induce oxidative stress inside bacterial cells was examined by measuring the levels of the antioxidant enzymes GPx and CAT. The amount of GPx significantly increased after treating the bacteria with N-SNPs and AgNO3. However, N-SNPs treatment caused the greatest increase in bacterial GPx. Among the examined bacterial species, MRSA treated with N-SNPs had the highest level of GPx. Additionally, GPx levels were unchanged in Salmonella typhimurium after treatment with AgNO3. The amount of CAT significantly decreased in all tested bacterial strains after treatment with 1.5 mg/mL of N-SNPs and AgNO3. N-SNP treatment caused the greatest reduction in CAT levels compared with control or AgNO3 treatments in all bacteria examined (Figure 4C and D).
Morphological Changes in E. coli Caused by N-SNPs and AgNO3
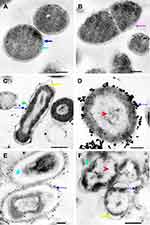
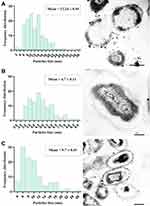
N-SNPs- and AgNO3-induced disruption of microbial cells was evaluated by TEM analysis. TEM micrographs of E. coli that had not been exposed to either silver species revealed intact, multi-layer, cell membranes comprising the outer membrane, a peptidoglycan layer in periplasmic space, and a cytoplasmic membrane (Figure 5A and B). In contrast, E. coli treated with 1.5 mg/mL AgNO3 for 24 h displayed several morphological changes including disintegration of cell membranes, the formation of pores and folds, detachment of the cell wall from the outer membrane, and the appearance of low-density areas in the center of the bacterial cells, suggestive of intensive cytoplasm disruption (Figure 5C and D). Moreover, the bacteria were slightly larger after AgNO3 treatment compared with untreated bacteria (Figure 6). Dark, dense electron granules, believed to be Ag-NPs produced from AgNO3 reduction by E. coli, were also adsorbed on the cellular membranes. These NPs were concentrated on the bacterial cell walls and the extracellular matrix, with a very small amount observed inside the bacterial cells. The average nanosize of E. coli-synthesized Ag-NPs was 12.24 ± 0.45 nm (Figure 7).
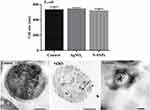 |
Figure 6 Size of E. coli before and after exposure to N-SNPs and AgNO3. Measurements were performed using ImageJ software based on TEM micrographs (at least ten cells). Scale bar: 200 nm. |
Ultrastructural changes were observed in E. coli cells after treatment with 1.5 mg/mL N-SNPs for 24 h (Figure 5E and F). Cell membranes of treated bacteria were shrunken, folded, and detached from the cell wall. Furthermore, intensive bacterial cytoplasmic lashing and nucleoagglutination were detected in all treated cells. E. coli exposed to N-SNPs were smaller than untreated bacteria (Figure 6). Dark, dense electron granules, believed to be N-SNPs, were adsorbed on the cellular membranes (Figure 8). N-SNPs were distributed throughout the cytosol of almost all treated cells, but were only observed on the bacterial cell walls of the more damaged cells. The average nanodiameter of cytosolic N-SNPs was 4.7 ± 0.13 nm, and that of N-SNPs adsorbed onto the cell walls was 9.7 ± 0.41 nm (Figure 7).
 |
Figure 8 TEM micrographs showing N-SNPs (gold arrow) precipitated on E. coli surface (A and B). Scale bars: 200 nm and 100 nm. |
Effect of N-SNPs and AgNO3 on Gene Expression in E. coli
The ability of N-SNPs and AgNO3 to influence expression of the genes mfD, flu, and hly in E. coli was examined using qRT-PCR (Figure 9). Both N-SNPs and AgNO3 significantly upregulated expression of mfD and significantly downregulated expression of flu and hly. N-SNP caused the most significant increase in mfD expression and the most significant decrease in flu and hly expression.
Effect of N-SNPs and AgNO3 on Protein Expression in E. coli
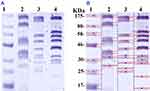
SDS-PAGE was used to study the effect of N-SNPs and AgNO3 on protein expression in E. coli (Figure 10). The protein profile of bacteria treated with AgNO3 revealed the presence of thinner and higher molecular weight (Mwt) protein bands, which were absent in untreated cells and in cells treated with N-SNPs. The protein profile of E. coli treated with N-SNPs contained lower Mwt protein bands that were absent in AgNO3-treated and untreated cells. In addition, more protein bands were observed in bacteria exposed to N-SNPs or AgNO3 than in control cells (10 bands in treated cells and six in control cells) (Table 4).
 |
Table 4 Number and Intensity of Protein Bands from E. coli Treated with and without N-SNPs and AgNO3 |
Discussion
N-SNPs exhibited greater antibacterial efficiency than AgNO3 against Gram-positive and Gram-negative bacteria. Similarly, determination of the MIC and MBC values revealed that lower concentrations of N-SNPs were needed to suppress bacterial growth compared with the concentrations of AgNO3. Among all tested bacteria, E. coli (IZ diameter of 18.6 ± 0.07 mm) and MRSA (IZ diameter of 18 ± 0.09 mm) were most sensitive to N-SNPs. These data suggest that N-SNPs are a potent antibacterial agent against both Gram-negative and Gram-positive bacteria. The slight increase in the IZ diameter observed for Gram-negative bacteria, compared with that of Gram-positive bacteria, may be the result of differences in bacterial cell-wall structures and the nano/cell interface pattern.51,59 Hamida et al reported that 1.5 mg/mL Ag-NPs, synthesized by Desertifilum sp. IPPAS B-1220, had inhibitory potential against different pathogenic bacteria (MRSA > Salmonella typhimurium > Klebsiella pneumoniae > E. coli > Streptococcus mutans),12 and that Ag-NPs were more effective against Gram-positive bacteria than against Gram-negative bacteria. The IZ diameter observed by Hamida et al after treating MRSA with Ag-NPs was 23.7 ± 0.08 mm while that of E. coli was 14.8 ± 0.07 mm. The IZ diameter for E. coli was smaller than that observed in the current study, implying that the antibacterial activity of Ag-NPs formed by Nostoc sp. Bahar_M is more effective than that of Ag-NPs synthesized by Desertifilum sp., at least against E. coli. This suggests that the coated functional groups (derived from different cyanobacterial strains) on the surface of NPs may affect the bacterial inhibitory activity of the NPs.20
The mode of action of Ag-NPs against microbes has not yet been elucidated. However, the lethal effect of Ag-NPs is thought to result from the release of Ag+ from Ag-NPs. Free Ag+ then interacts with several cellular sites including membranes, the cytoplasm, and the nuclear matrix. The nano/cell interface interactions result in loss of K+ from the cellular membranes, increased membrane permeability, impaired membrane integrity, and effects on the respiration chain and biomolecules including DNA, proteins, and enzymes.60,61 Other reports suggest that toxicity of Ag-NP is related to its ability to induce oxidative stress by stimulating the formation of ROS, which results in disruption of cellular biomolecules and structures leading to cellular dysfunction and cell death.31,35,57
LDH, ATPase, and antioxidant (GPx and CAT) enzyme analyses revealed that N-SNPs were more effective than AgNO3 as an antibacterial agent, and disrupted bacterial enzyme activities. LDH is a marker of cell death and membrane rupture.62,63 The significant increase in LDH levels after treating the bacteria with N-SNPs and AgNO3 indicates that both silver species affected the integrity and permeability of bacterial membranes. The increase in LDH after treating bacteria with N-SNPs was greater than that observed for AgNO3, suggesting that silver, in the nanoform (N-SNPs), has greater toxicity than silver ion (AgNO3).54 This may be because of the unique features of NPs including their smaller size to larger surface area, their surface chemistry, and their charges that enable them to easily attach and penetrate bacterial cell walls and membranes.58 These results are consistent with those of Korshed et al who reported that LDH levels in E. coli increased in a dose-dependent manner after exposure to laser Ag-NPs, suggesting their potential to increase membrane permeability and protein leakage, leading to cell death.64 Hamida et al demonstrated that treatment with Ag-NPs, synthesized by Nostoc sp., and AgNO3 significantly increased LDH levels in K. pneumoniae, and that Ag-NPs caused a more significant surge in enzyme activity compared with AgNO3.20
With exception of Streptococcus mutans, the significant decrease in ATPase observed after treating bacterial cells with N-SNPs and AgNO3 could be attributed to the ability of these treatments to induce metabolic toxicity by directly interacting with ATPase or by enhancing oxidative stress resulting in ATPase denaturation and dysfunction. Cui et al reported that gold NPs caused metabolic toxicity in E. coli by interacting with ATPase.65 Similarly, Ag-NPs synthesized using Desertifilum sp. significantly decreased the ATPase activity of MRSA, E. coli, K. pneumoniae, Streptococcus mutans, and Salmonella typhimurium.54 In the current study, the greater decrease in ATPase levels observed after N-SNP treatment compared with AgNO3 treatment may be due to the smaller size of N-SNPs and their surface chemistry.66 In contrast, the significant increase in ATPase level in Streptococcus mutans after treatment with N-SNPs might be attributed to the ability of these bacteria to resist N-SNPs by producing more energy. This result was consistent with the findings of the agar well diffusion assay in which Streptococcus mutans had the weakest response to N-SNPs (IZ diameter of 14.7 ± 0.03 mm) of all bacteria tested. Congruent with these data, Wang et al reported that copper-NPs increased ATPase activity in juvenile Epinephelus coioides.67
N-SNPs caused significantly larger decreases in CAT and increases in GPx activities compared with AgNO3, indicating that N-SNPs have a greater potential than AgNO3 to stimulate ROS production, leading cellular oxidative stress induction.12,20,68
TEM analysis of E. coli before and after exposure to N-SNPs and AgNO3 revealed that both silver species caused marked ultrastructural changes in the bacteria. However, N-SNP treatment resulted in the most acute morphological changes. These cellular alterations may be because of N-SNPs/cell interface or indirect influence through enhancing oxidative stress by NPs, resulting in bacterial death.9,69 The ultrastructural changes observed in E. coli after treatment with N-SNPs had two distinct patterns. The first included changes in cellular borders including irregularity, disruption of bacterial cell wall and membranes, folded and pored membranes, multilayered membrane shrinkage, and detachment of the cellular membrane from the cytoplasmic matrix.9,51 These changes were consistent with data from the LDH assays in which N-SNPs caused a significant surge in LDH activity in all tested bacteria, indicating that N-SNPs negatively impact bacterial membrane integrity and permeability.
Moreover, N-SNPs were observed on the surface of bacteria and were also concentrated inside the bacterial cytoplasm. N-SNPs adsorbed on bacterial membranes had an average nanosize of 9.7 ± 0.41 nm, while N-SNPs concentrated in the cytoplasm had an average nanodiameter of 4.7 ± 0.13 nm. The larger size of N-SNPs outside bacterial cells than inside the cells may be due to agglomeration of NPs outside the cells allowing only smaller-sized particles to pass inside the cells.58 We hypothesize that N-SNPs are electrostatically attracted to cell membranes then adsorbed on the bacterial surfaces causing membrane folds and pores as well as changes in membrane permeability and integrity.51,70,71 These steps facilitate entry of N-SNPs into bacterial cells where they interact with cytoplasmic and nucleic contents, leading to bacterial cell death. Xu et al reported that Ag-NPs decorated with carboxylate (higher oxidation degree) or hydroxylate (lower oxidation capacity) carbon dots (C-CDs and H-CDs, respectively) significantly inhibited growth of E. coli and Staphylococcus aureus.51 However, the effect of these nanoagents was greater in Gram-negative bacteria than in Gram-positive bacteria. A concentration of 0.4 nmol/L Ag-NPs@H-CDs and Ag-NPs@C-CDs caused antibacterial efficiencies of 99.9% and 76.4%, respectively, in E. coli, while in Staphylococcus aureus 0.8 nmol/L of both nanoagents was required to achieve antibacterial efficiencies of 99.9% and 86.5%, respectively. The authors suggested that differences in antibacterial efficacies of the nanodrugs against both types of bacteria were due to nano/cell interface interactions.
The second pattern of ultrastructural changes in the bacteria treated with N-SNPs comprised morphological alterations in cytoplasmic and nuclear contents, and included severe cytoplasmic lashing and nucleoagglomeration. These data indicate that N-SNPs may enhance bacterial apoptosis by directly interacting with cytoplasmic and nucleic contents including proteins, enzymes, and nucleic acids, or by inducing ROS formation, which results in intensive oxidative stress and DNA damage, biomolecule denaturation, and cell damage12,55 (Figure 11).
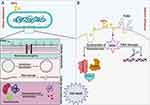 |
Figure 11 Schematic diagram illustrating direct (A) and indirect (B) influences of NPs on bacterial cells. Created by Biorender. |
In addition, the TEM micrographs of E. coli treated with the silver nitrate showed small dark granules with an average nanosize of 12.24 ± 0.45 nm, indicating that E. coli had the potential to reduce AgNO3 into Ag-NPs. We suggest that the Ag-NPs synthesized by E. coli were responsible for the observed bacterial cellular alterations. Indeed, E. coli treated with AgNO3 showed similar changes to those caused by treatment with N-SNPs, including membrane disruption, folded and pored membranes, detachment of bacterial membranes, and moderate cytoplasmic dissolution. These observations were consistent with those of Jung et al who utilized TEM analysis to detect ultrastructural changes in E. coli and Staphylococcus aureus before and after AgNO3 treatment.72 Additionally, El-Shanshoury reported that E. coli could reduce AgNO3 into Ag-NPs,73 and that Ag-NPs fabricated by E. coli ranged from 5 nm to 25 nm in size. However, N-SNPs caused more severe ultrastructural changes in the bacteria than Ag-NPs synthesized by E. coli. This may be due to the smaller size and surface chemistry (Nostoc biomolecule coating) of N-SNPs, making them more potent (Figure 12).
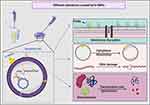 |
Figure 12 Schematic diagram showing different structural and biomolecule alterations in bacterial cells caused by N-SNPs. Created using Biorender. |
To understand the influence of N-SNPs and AgNO3 on E. coli pathogenicity, the expression of different genes related to DNA repair (mfD), biofilm formation (flu), and virulence activity (hly) was examined. Treatment with N-SNPs and AgNO3 caused changes in the expression levels of these genes in E. coli. N-SNP treatment resulted in the most significant changes in expression levels of the tested genes. This may because of the smaller size of the NPs and the functional groups on the NP surface (derived from Nostoc sp.) enabling them to interact with DNA.66 The significant increase in mfD expression indicates that N-SNPs lead to DNA structural disruption by direct interaction with DNA molecules or/and indirectly by enhancing oxidative stress.74,75 Similarly, downregulation of both flu and hly suggests that N-SNPs have the potential to inhibit biofilm formation and bacterial invasion via direct or/and indirect interference with bacterial genes.54,74,76
Protein profiles of E. coli treated with N-SNPs and AgNO3 were examined by SDS-PAGE. Exposure to both silver species led to the appearance of new protein bands (10 bands), indicating that both silver species enhance protein denaturation and degradation comparing to the control (6 bands). E. coli treated with N-SNPs had lower Mwt protein bands than those treated with AgNO3, suggesting that bacterial proteins are more sensitive to N-SNPs. The greater toxicity of N-SNPs toward bacterial proteins may again be a result of their unique physicochemical features such as smaller size and surface chemistry. These properties may increase the affinity between NPs and cellular biomolecules, resulting in protein chain unfolding, and potentially leading to protein modification and degradation.54,77 Conversely, the appearance of extra protein bands may indicate the formation of new proteins by the bacteria as a defense system against both silver species.78,79
Upon the previous data, we speculated that the killing mechanisms of N-SNPs and AgNO3 against E. coli may be followed two predominant strategies. The first is the nano/cell interface in which NPs directly interfere with cell wall and cell membranes, resulting in disruption of membrane permeability and integrity through the formation of folds and pores. This process facilitates entry of NPs into cells to interact with and damage cellular biomolecules including DNA, proteins, and enzymes, leading to dysfunction and bacterial cell death. The second strategy is the induction of ROS formation by NPs, which causes intensive oxidative stress and results in biomolecule dysfunction, structural damage to cells, and ultimately apoptosis (Figure 13). These results demonstrate that ecofriendly N-SNPs may serve as potent antibacterial agents against pathogenic and MDR bacteria through a plethora of effects on bacteria. These effects include increased membrane permeability and disruption; disturbance of bacterial biological functions such as biofilm formation and virulence activity; induction of oxidative stress; generation of genetic damage; and interference with biomolecules causing cellular dysfunction.
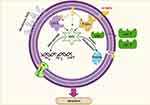 |
Figure 13 Diagram demonstrating the potential mechanism of action of N-SNPs against E. coli. Created using Biorender. |
Conclusion
In this study, the inhibitory effect of Ag-NPs, synthesized by Nostoc sp. Bahar_M., and AgNO3 against four pathogenic bacteria were examined for first time. In addition, the mechanism of N-SNPs- and AgNO3-mediated lethality against E. coli was explained based on molecular and morphological analyses. Both silver species inhibited bacterial growth, increase the leakage of LDH, depletion ATPase activity and imbalance antioxidants (CAT and GPx) activity leading to enhance the oxidative stress and metabolic toxicity. They caused imbalance in mfD, flu, hly genes expression and protein degradation. However, N-SNPs were more effective than AgNO3 against all tested bacteria. The possible mechanisms of N-SNPs- and AgNO3-mediated lethality against E. coli were including direct nano/cell interface via interfering with cellular structures and/or via indirect influence through stimulating the production of ROS which causes intensive oxidative stress and results in biomolecule dysfunction, structural damage to cells, and ultimately apoptosis.
Further research is required to investigate the inhibitory activity of N-SNPs against other pathogenic bacteria, with an emphasis on elucidating the molecular mechanisms of action of these NPs against bacterial cells. Moreover, studies on the synergetic effects of N-SNPs in combination with other antibiotics are necessary to determine the ability of these NPs to improve antibiotic activity against MDR bacteria.
Abbreviations
Ag-NPs, silver nanoparticles; NPs, nanoparticles; µL, microliter; h, hour; mL, milliliter; rpm, revolutions per minute; kb, kilobase; min, minute; mg, milligram; mM, millimolar; µg, microgram; Mwt, molecular weight.
Data Sharing Statement
Data supporting this article are shown in Figures 1–13 and Tables 1–4. The data sets analyzed in the present study are available from the corresponding author upon reasonable request.
Ethics Statement
The current study was approved by the Research Ethics committee of Faculty of Medicine, Alexandria University, Alexandria, Egypt, granted permission for this work (IRB NO: 00012098).
Acknowledgments
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest.
References
1. Dye C. After 2015: infectious diseases in a new era of health and development. Philos Trans R Soc B Biol Sci. 2014;369(1645):20130426. doi:10.1098/rstb.2013.0426
2. Fonkwo PN. Pricing infectious disease: the economic and health implications of infectious diseases. EMBO Rep. 2008;9(S1):S13–S17. doi:10.1038/embor.2008.110
3. Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42–51.
4. Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303(6–7):287–292. doi:10.1016/j.ijmm.2013.02.009
5. Murphy CN, Clegg S. Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol. 2012;7(8):991–1002. doi:10.2217/fmb.12.74
6. Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482. doi:10.3934/microbiol.2018.3.482
7. Guillard T, Pons S, Roux D, Pier GB, Skurnik D. Antibiotic resistance and virulence: understanding the link and its consequences for prophylaxis and therapy. Bioessays. 2016;38(7):682–693. doi:10.1002/bies.201500180
8. Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9(2):117–128. doi:10.1038/nrd3013
9. Vazquez-Muñoz R, Meza-Villezcas A, Fournier P, et al. Enhancement of antibiotics antimicrobial activity due to the silver nanoparticles impact on the cell membrane. PLoS One. 2019;14(11):e0224904. doi:10.1371/journal.pone.0224904
10. Huh AJ, Kwon YJ. “Nanoantibiotics”: a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J Control Release. 2011;156(2):128–145. doi:10.1016/j.jconrel.2011.07.002
11. Weissig V, Pettinger TK, Murdock N. Nanopharmaceuticals (part 1): products on the market. Int J Nanomedicine. 2014;9:4357. doi:10.2147/IJN.S46900
12. Hamida RS, Ali MA, Goda DA, Khalil MI, Al-Zaban MI. Novel biogenic silver nanoparticle-induced reactive oxygen species inhibit the biofilm formation and virulence activities of Methicillin-Resistant Staphylococcus aureus (MRSA) strain. Front Bioeng Biotechnol. 2020;8:1–14. doi:10.3389/fbioe.2020.00433
13. Das S, Ranjana N, Misra AJ, et al. Disinfection of the water borne pathogens Escherichia coli and Staphylococcus aureus by solar photocatalysis using sonochemically synthesized reusable Ag@ ZnO core-shell nanoparticles. Int J Environ Res Public Health. 2017;14(7):747. doi:10.3390/ijerph14070747
14. Sheel R, Kumari P, Panda PK, et al. Molecular intrinsic proximal interaction infer oxidative stress and apoptosis modulated in vivo biocompatibility of P. niruri contrived antibacterial iron oxide nanoparticles with zebrafish. Environ Pollut. 2020;267:115482. doi:10.1016/j.envpol.2020.115482
15. Mazurak VC, Burrell RE, Tredget EE, Clandinin MT, Field CJ. The effect of treating infected skin grafts with Acticoat™ on immune cells. Burns. 2007;33(1):52–58. doi:10.1016/j.burns.2006.04.027
16. Bobo D, Robinson KJ, Islam J, Thurecht KJ, Corrie SR. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm Res. 2016;33(10):2373–2387.
17. Hamida RS, Ali MA, Redhwan A, Bin-Meferij MM. Cyanobacteria–a promising platform in green nanotechnology: a review on nanoparticles fabrication and their prospective applications. Int J Nanomedicine. 2020;15:6033–6066.
18. da Silva BL, Abuçafy MP, Manaia EB, et al. Relationship between structure and antimicrobial activity of zinc oxide nanoparticles: an overview. Int J Nanomedicine. 2019;14:9395. doi:10.2147/IJN.S216204
19. Verma SK, Jha E, Kiran K, Bhat S, Suar M, Mohanty P. Synthesis and characterization of novel polymer-hybrid silver nanoparticles and its biomedical study. Mater Today. 2016;3(6):1949–1957.
20. Hamida RS, Ali MA, Goda DA, Khalil MI, Redhwan A. Cytotoxic effect of green silver nanoparticles against ampicillin-resistant Klebsiella pneumoniae. RSC Adv. 2020;10(36):21136–21146. doi:10.1039/D0RA03580G
21. Kumari S, Kumari P, Panda PK, et al. Biocompatible biogenic silver nanoparticles interact with caspases on an atomic level to elicit apoptosis. Nanomedicine. 2020;15(22):2119–2132.
22. Pannerselvam B, Alagumuthu TS, Cinnaiyan SK, et al. In vitro cytotoxicity and antibacterial activity of optimized silver nanoparticles against wound infectious bacteria and their morphological studies. J Cluster Sci. 2020;1–14.
23. Iravani S, Korbekandi H, Mirmohammadi SV, Zolfaghari B. Synthesis of silver nanoparticles: chemical, physical and biological methods. Res Pharm Sci. 2014;9(6):385.
24. Verma SK, Nisha K, Panda PK, et al. Green synthesized MgO nanoparticles infer biocompatibility by reducing in vivo molecular nanotoxicity in embryonic zebrafish through arginine interaction elicited apoptosis. Sci Total Environ. 2020;713:136521. doi:10.1016/j.scitotenv.2020.136521
25. Verma SK, Jha E, Panda PK, et al. Rapid novel facile biosynthesized silver nanoparticles from bacterial release induce biogenicity and concentration dependent in vivo cytotoxicity with embryonic zebrafish—a mechanistic insight. Toxicol Sci. 2018;161(1):125–138. doi:10.1093/toxsci/kfx204
26. Hamida RS, Abdelmeguid NE, Ali MA, Bin-Meferij MM, Khalil MI. Synthesis of silver nanoparticles using a novel cyanobacteria Desertifilum sp. extract: their antibacterial and cytotoxicity effects. Int J Nanomedicine. 2020;15:49. doi:10.2147/IJN.S238575
27. Bin-Meferij MM, Hamida RS. Biofabrication and antitumor activity of silver nanoparticles utilizing novel nostoc sp. Bahar M. Int J Nanomedicine. 2019;14:9019. doi:10.2147/IJN.S230457
28. Wei L, Lu J, Xu H, Patel A, Chen Z-S, Chen G. Silver nanoparticles: synthesis, properties, and therapeutic applications. Drug Discov Today. 2015;20(5):595–601. doi:10.1016/j.drudis.2014.11.014
29. Zhang XF, Liu ZG, Shen W, Gurunathan S. Silver nanoparticles: synthesis, characterization, properties, applications, and therapeutic approaches. Int J Mol Sci. 2016;17(9):1534.
30. Patel V, Berthold D, Puranik P, Gantar M. Screening of cyanobacteria and microalgae for their ability to synthesize silver nanoparticles with antibacterial activity. Biotechnol Rep. 2015;5:112–119. doi:10.1016/j.btre.2014.12.001
31. Hamouda RA, Hussein MH, Abo-elmagd RA, Bawazir SS. Synthesis and biological characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica. Sci Rep. 2019;9(1):1–17.
32. Paul P, Verma S, Kumar Panda P, Jaiswal S, Sahu BR, Suar M. Molecular insight to influential role of Hha–TomB toxin–antitoxin system for antibacterial activity of biogenic silver nanoparticles. Artif Cells, Nanomed Biotechnol. 2018;46(sup3):S572–S584. doi:10.1080/21691401.2018.1503598
33. Kumari S, Panda PK, Pramanik N, Verma SK, Mallick M. Molecular aspect of phytofabrication of gold nanoparticle from Andrographis peniculata photosystem II and their in vivo biological effect on embryonic zebrafish (Danio rerio). Environ Nanotechnol Monit Manag. 2019;11:100201. doi:10.1016/j.enmm.2018.100201
34. Misra AJ, Das S, Rahman AH, et al. Doped ZnO nanoparticles impregnated on Kaolinite (Clay): a reusable nanocomposite for photocatalytic disinfection of multidrug resistant Enterobacter sp. under visible light. J Colloid Interface Sci. 2018;530:610–623. doi:10.1016/j.jcis.2018.07.020
35. Dakal TC, Kumar A, Majumdar RS, Yadav V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front Microbiol. 2016;7:1831.
36. Murugesan K, Koroth J, Srinivasan PP, et al. Effects of green synthesised silver nanoparticles (ST06-AgNPs) using curcumin derivative (ST06) on human cervical cancer cells (HeLa) in vitro and EAC tumor bearing mice models. Int J Nanomedicine. 2019;14:5257. doi:10.2147/IJN.S202404
37. Verma SK, Jha E, Panda PK, et al. Mechanistic insight into size-dependent enhanced cytotoxicity of industrial antibacterial titanium oxide nanoparticles on colon cells because of reactive oxygen species quenching and neutral lipid alteration. ACS Omega. 2018;3(1):1244–1262. doi:10.1021/acsomega.7b01522
38. Verma SK, Jha E, Panda PK, et al. Molecular insights to alkaline based bio-fabrication of silver nanoparticles for inverse cytotoxicity and enhanced antibacterial activity. Mater Sci Eng C. 2018;92:807–818. doi:10.1016/j.msec.2018.07.037
39. Kanagamani K, Muthukrishnan P, Shankar K, Kathiresan A, Barabadi H, Saravanan M. Antimicrobial, cytotoxicity and photocatalytic degradation of norfloxacin using Kleinia grandiflora mediated silver nanoparticles. J Cluster Sci. 2019;30(6):1415–1424. doi:10.1007/s10876-019-01583-y
40. Jebril S, Jenana RKB, Dridi C. Green synthesis of silver nanoparticles using melia azedarach leaf extract and their antifungal activities: in vitro and in vivo. Mater Chem Phys. 2020;248:122898. doi:10.1016/j.matchemphys.2020.122898
41. Amarasinghe L, Wickramarachchi P, Aberathna A, Sithara W, De Silva C. Comparative study on larvicidal activity of green synthesized silver nanoparticles and Annona glabra (Annonaceae) aqueous extract to control Aedes aegypti and Aedes albopictus (Diptera: culicidae). Heliyon. 2020;6(6):e04322. doi:10.1016/j.heliyon.2020.e04322
42. Mori Y, Ono T, Miyahira Y, Nguyen VQ, Matsui T, Ishihara M. Antiviral activity of silver nanoparticle/chitosan composites against H1N1 influenza A virus. Nanoscale Res Lett. 2013;8(1):1–6. doi:10.1186/1556-276X-8-93
43. Hamida RS, Albasher G, Bin-Meferij MM. Oxidative stress and apoptotic responses elicited by nostoc-synthesized silver nanoparticles against different cancer cell lines. Cancers. 2020;12(8):2099.
44. Jyoti K, Arora D, Fekete G, Lendvai L, Dogossy G, Singh T. Antibacterial and anti-inflammatory activities of Cassia fistula fungal broth-capped silver nanoparticles. Mater Technol. 2020;1–11. doi:10.1080/10667857.2020.1802841
45. Junejo Y, Safdar M, Akhtar MA, et al. Synthesis of tobramycin stabilized silver nanoparticles and its catalytic and antibacterial activity against pathogenic bacteria. J Inorg Organomet Polym Mater. 2019;29(1):111–120. doi:10.1007/s10904-018-0971-z
46. Durán N, Durán M, De Jesus MB, Seabra AB, Fávaro WJ, Nakazato G. Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity. Nanomedicine. 2016;12(3):789–799. doi:10.1016/j.nano.2015.11.016
47. Ghiuță I, Cristea D. Silver nanoparticles for delivery purposes. Nanoengineered Biomater Adv Drug Deliv. 2020;347.
48. Abo-Shama UH, El-Gendy H, Mousa WS, et al. Synergistic and antagonistic effects of metal nanoparticles in combination with antibiotics against some reference strains of pathogenic microorganisms. Infect Drug Resist. 2020;13:351. doi:10.2147/IDR.S234425
49. Ruden S, Hilpert K, Berditsch M, Wadhwani P, Ulrich AS. Synergistic interaction between silver nanoparticles and membrane-permeabilizing antimicrobial peptides. Antimicrob Agents Chemother. 2009;53(8):3538–3540. doi:10.1128/AAC.01106-08
50. Cavassin ED, de Figueiredo LFP, Otoch JP, et al. Comparison of methods to detect the in vitro activity of silver nanoparticles (AgNP) against multidrug resistant bacteria. J Nanobiotechnology. 2015;13(1):64. doi:10.1186/s12951-015-0120-6
51. Xu Z, He H, Zhang S, et al. Mechanistic studies on the antibacterial behavior of Ag nanoparticles decorated with carbon dots having different oxidation degrees. Environ Sci. 2019;6(4):1168–1179.
52. Yin IX, Zhang J, Zhao IS, Mei ML, Li Q, Chu CH. The antibacterial mechanism of silver nanoparticles and its application in dentistry. Int J Nanomedicine. 2020;15:2555. doi:10.2147/IJN.S246764
53. Ramkumar VS, Pugazhendhi A, Gopalakrishnan K, et al. Biofabrication and characterization of silver nanoparticles using aqueous extract of seaweed Enteromorpha compressa and its biomedical properties. Biotechnol Rep. 2017;14:1–7. doi:10.1016/j.btre.2017.02.001
54. Hamida RSAMA, Goda AD, Redhwan AO, Khalil MI. Antibacterial, anti-virulence and anti-biofilm formation activities of biofabricated silver nanoparticles against drug-resistant Klebsiella pneumonia. Int J Nanomedicine. 2020;1–19.
55. Yuan Y-G, Peng Q-L, Gurunathan S. Effects of silver nanoparticles on multiple drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa from mastitis-infected goats: an alternative approach for antimicrobial therapy. Int J Mol Sci. 2017;18(3):569–591. doi:10.3390/ijms18030569
56. Andrés MT, Fierro JF. Antimicrobial mechanism of action of transferrins: selective inhibition of H+-ATPase. Antimicrob Agents Chemother. 2010;54(10):4335–4342. doi:10.1128/AAC.01620-09
57. Gurunathan S, Qasim M, Park C, et al. Cytotoxicity and transcriptomic analysis of silver nanoparticles in mouse embryonic fibroblast cells. Int J Mol Sci. 2018;19(11):3618–3641. doi:10.3390/ijms19113618
58. Romero-Urbina DG, Lara HH, Velázquez-Salazar JJ, et al. Ultrastructural changes in methicillin-resistant Staphylococcus aureus induced by positively charged silver nanoparticles. Beilstein J Nanotechnol. 2015;6(1):2396–2405. doi:10.3762/bjnano.6.246
59. Huq M. Green synthesis of silver nanoparticles using Pseudoduganella eburnea MAHUQ-39 and their antimicrobial mechanisms investigation against drug resistant human pathogens. Int J Mol Sci. 2020;21(4):1510. doi:10.3390/ijms21041510
60. Xiu Z-M, Ma J, Alvarez PJJ. Differential effect of common ligands and molecular oxygen on antimicrobial activity of silver nanoparticles versus silver ions. Environ Sci Technol. 2011;45(20):9003–9008. doi:10.1021/es201918f
61. Ivask A, ElBadawy A, Kaweeteerawat C, et al. Toxicity mechanisms in Escherichia coli vary for silver nanoparticles and differ from ionic silver. ACS Nano. 2014;8(1):374–386. doi:10.1021/nn4044047
62. Chan FK-M, Moriwaki K, De Rosa MJ. Detection of necrosis by release of lactate dehydrogenase activity. In: Snow A, Lenardo M, editors. Immune Homeostasis. Methods in Molecular Biology (Methods and Protocols). Totowa, NJ: Springer; 2013;979:65–70. doi:10.1007/978-1-62703-290-2_7
63. Burd J, Usategui-Gomez M. A colorimetric assay for serum lactate dehydrogenase. Clin Chim Acta. 1973;46(3):223–227. doi:10.1016/0009-8981(73)90174-5
64. Korshed P, Li L, Liu Z, Wang T, Gan Y. The molecular mechanisms of the antibacterial effect of picosecond laser generated silver nanoparticles and their toxicity to human cells. PLoS One. 2016;11(8):e0160078. doi:10.1371/journal.pone.0160078
65. Cui Y, Zhao Y, Tian Y, Zhang W, Lü X, Jiang X. The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials. 2012;33(7):2327–2333. doi:10.1016/j.biomaterials.2011.11.057
66. Tang S, Zheng J. Antibacterial activity of silver nanoparticles: structural effects. Adv Healthcare Mater. 2018;7(13):1701503–1701513. doi:10.1002/adhm.201701503
67. Wang T, Long X, Cheng Y, Liu Z, Yan S. The potential toxicity of copper nanoparticles and copper sulphate on juvenile Epinephelus coioides. Aquat Toxicol. 2014;152:96–104. doi:10.1016/j.aquatox.2014.03.023
68. Gurunathan S, Choi Y-J, Kim J-H. Antibacterial efficacy of silver nanoparticles on endometritis caused by Prevotella melaninogenica and Arcanobacterum pyogenes in dairy cattle. Int J Mol Sci. 2018;19(4):1210. doi:10.3390/ijms19041210
69. Choi O, Hu Z. Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria. Environ Sci Technol. 2008;42(12):4583–4588. doi:10.1021/es703238h
70. Li W-R, Xie X-B, Shi Q-S, Zeng H-Y, OU-Yang Y-S, Chen Y-B. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl Microbiol Biotechnol. 2010;85(4):1115–1122. doi:10.1007/s00253-009-2159-5
71. Badwaik VD, Vangala LM, Pender DS, et al. Size-dependent antimicrobial properties of sugar-encapsulated gold nanoparticles synthesized by a green method. Nanoscale Res Lett. 2012;7(1):1–11. doi:10.1186/1556-276X-7-623
72. Jung WK, Koo HC, Kim KW, Shin S, Kim SH, Park YH. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl Environ Microbiol. 2008;74(7):2171–2178. doi:10.1128/AEM.02001-07
73. El-Shanshoury AE-R-R, ElSilk SE, Ebeid ME. Extracellular biosynthesis of silver nanoparticles using Escherichia coli ATCC 8739, Bacillus subtilis ATCC 6633, and Streptococcus thermophilus ESh1 and their antimicrobial activities. ISRN Nanomater. 2011;2011:1–7. doi:10.5402/2011/385480
74. Radzig M, Nadtochenko V, Koksharova O, Kiwi J, Lipasova V, Khmel I. Antibacterial effects of silver nanoparticles on gram-negative bacteria: influence on the growth and biofilms formation, mechanisms of action. Colloids Surf B Biointerfaces. 2013;102:300–306. doi:10.1016/j.colsurfb.2012.07.039
75. Ashmore DA, Chaudhari A, Barlow B, et al. Evaluation of E. coli inhibition by plain and polymer-coated silver nanoparticles. Rev Inst Med Trop Sao Paulo. 2018;60:1–11. doi:10.1590/s1678-9946201860018
76. Vishnupriya S, Chaudhari K, Jagannathan R, Pradeep T. Single‐cell investigations of silver nanoparticle–bacteria interactions. Part Part Syst Charact. 2013;30(12):1056–1062.
77. Soliman H, Elsayed A, Dyaa A. Antimicrobial activity of silver nanoparticles biosynthesised by Rhodotorula sp. strain ATL72. Egypt J Basic Appl Sci. 2018;5(3):228–233. doi:10.1016/j.ejbas.2018.05.005
78. Gogoi SK, Gopinath P, Paul A, Ramesh A, Ghosh SS, Chattopadhyay A. Green fluorescent protein-expressing Escherichia coli as a model system for investigating the antimicrobial activities of silver nanoparticles. Langmuir. 2006;22(22):9322–9330. doi:10.1021/la060661v
79. Velusamy P, Kumar GV, Jeyanthi V, Das J, Pachaiappan R. Bio-inspired green nanoparticles: synthesis, mechanism, and antibacterial application. Toxicol Res. 2016;32(2):95–102. doi:10.5487/TR.2016.32.2.095
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.