Back to Journals » OncoTargets and Therapy » Volume 13
Knockdown of TRIM37 Promotes Apoptosis and Suppresses Tumor Growth in Gastric Cancer by Inactivation of the ERK1/2 Pathway
Authors Zhu H, Chen Y, Zhang J, Qian C, Qiu W, Shen H, Shen Z
Received 8 October 2019
Accepted for publication 20 May 2020
Published 12 June 2020 Volume 2020:13 Pages 5479—5491
DOI https://doi.org/10.2147/OTT.S233906
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Arseniy Yuzhalin
Hongyi Zhu,* Yuanwen Chen,* Jie Zhang, Changlin Qian, Weiqing Qiu, Huojian Shen, Zhiyong Shen
Department of General Surgery, South Campus, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 201112, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Huojian Shen; Zhiyong Shen
Department of General Surgery, South Campus, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, 2000 JiangYue Road, Minhang District, Shanghai 201112, People’s Republic of China
Tel +8621-58752345
Email [email protected]; [email protected]
Objective: Gastric cancer (GC), a malignant tumor of the gastric mucosa, is the second leading cause of cancer deaths worldwide. Although the incidence and mortality of gastric cancer have been reduced in the US and elsewhere, it is still a major public health concern. In this study, we attempted to investigate the function of tripartite motif-containing protein 37 (TRIM37) in GC cell lines in order to propose a new therapy for GC.
Methods: The expression of TRIM37 in GC patients and cell lines was detected by immunohistochemistry, real-time PCR and Western blotting analysis. After TRIM37 knockdown or overexpression, the cell cycle, proliferation and apoptosis, as well as the expression of related proteins, were detected. In addition, in vivo experiments on nude mice were performed.
Results: We found that TRIM37 expression was significantly elevated in tumor tissues of GC patients and GC cell lines, and patients with high expression of TRIM37 had a poor prognosis. Knockdown of TRIM37 in GC cells significantly inhibited cell proliferation and cell cycle progression, promoted apoptosis, increased cleaved caspase 3 and decreased c-myc and phosphorylation of protein kinase 1/2 (p-ERK1/2). Effects of TRIM37 overexpression were opposite to that of TRIM37 knockdown and were potently attenuated by an ERK1/2 inhibitor. In addition, an ERK1/2 agonist increased TRIM37 and p-ERK1/2 in a dose-dependent manner, and TRIM37 knockdown potently attenuated EGF-induced cell proliferation and expression of TRIM37 and p-ERK1/2. Interestingly, we found that TRIM37 overexpression did not affect the mRNA level of dual-specificity phosphatase 6 (DUSP6), but reduced its protein level in GC cells. Co-immunoprecipitation (Co-IP) analyses revealed that TRIM37 interacted with DUSP6, and TRIM37 overexpression enhanced DUSP6 ubiquitination in GC cells. In vivo experiments on nude mice showed the inhibitory effect of TRIM37 knockdown on tumor growth.
Conclusion: These findings suggest that TRIM37 may act as an oncogene in the growth of GC cells and illustrate its potential function as a target in the treatment of GC.
Keywords: TRIM37, gastric cancer, ERK1/2 pathway, cleaved caspase 3, C-myc
Introduction
Gastric cancer (GC), a malignant tumor of the gastric mucosa, is the second leading cause of cancer deaths worldwide.1–3 Data have shown that compared to the southern regions, the incidence of GC in the northwest and eastern coastal areas of China are much higher.4 GC not only causes damage to the digestive system, but metastasis of GC can affect liver, kidney and respiratory functions.5 In severe cases, GC may result in cachexia and is ultimately life-threatening. Currently, fiber endoscopy, supplemented with X-ray barium meal examination, abdominal B-ultrasound, tumor markers, routine blood examination, and gastric juice analysis is the most effective way to diagnose gastric cancer. In recent years, advances in diagnostic techniques have increased early detection of gastric cancer thereby reducing mortality, but recurrent disease often occurs in patients with advanced disease, although aggressively treated, their survival rate is extremely low.6
Tripartite motif-containing protein 37 (TRIM37), also known as MUL, contains a RING finger domain, and is identified to be an E3 ubiquitin ligase.7 TRIM37 is located on the 17q23 chromosome, which is reported to be amplified in up to 40% of breast cancers.8 As a carcinogenic H2A ubiquitin ligase, TRIM37 is overexpressed in breast cancer and can promote transformation by silencing tumor suppressors and other genes.9 Additionally, studies have reported that TRIM37 is associated with the growth, migration and metastasis in multiple cancers, such as pancreatic cancer, glioma, hepatocellular carcinoma and colorectal cancer.10–14 Furthermore, knockdown of TRIM37 can inhibit human glioma cell proliferation and metastasis via activation of the PI3K/AKT pathway, while in liver cancer, overexpression of TRIM37 can promote cell migration and metastasis through activation of the Wnt/β-catenin pathway.11,12 In this study, we attempted to explore the function and potential mechanism of TRIM37 in human GC cells to put forward a novel therapeutic target for GC treatment.
Dual-specificity phosphatase 6 (DUSP6), a cytosolic phosphatase, is a negative-feedback regulator for the extracellular-signal-regulated kinase 1/2 (ERK1/2).15 Studies have reported that DUSP6 plays a tumor suppressive role in lung cancer.16,17 DUSP6 has a neuroprotective effect on Aβ-induced cytotoxicity via suppression of ERK1/2 activation in neural stem cells, and down-regulation of DUSP6 contributes to the progression and differentiation of esophageal squamous cell carcinoma (ESCC).18,19 Furthermore, TRIM7 is reported to promote hepatocellular carcinoma cell proliferation via the DUSP6/p38 pathway, and TRIM11 downregulation inhibited DUSP6 protein in D-54 glioblastoma multiforme (GBM) cells.20,21 However, the interaction between TRIM37 and DUSP6 in GC cells has not been elucidated.
Here, elevated TRIM37 expression was observed in GC tumor tissues and cell lines, and elevated expression of TRIM37 in patients correlated with a poor prognosis. Knockdown of TRIM37 in GC cells significantly inhibited cell proliferation and cycle progression, promoted apoptosis, increased cleaved caspase 3 and decreased c-myc and phosphorylated ERK1/2 (p-ERK1/2). Effects of TRIM37 overexpression were opposite to TRIM37 knockdown and was potently attenuated by an ERK1/2 inhibitor, U0126. In addition, ERK1/2 activator, EGF, increased TRIM37 and p-ERK1/2 levels in a dose-dependent manner, while TRIM37 knockdown potently attenuated EGF-induced cell proliferation and expression of TRIM37 and p-ERK1/2. Co-Immunoprecipitation (Co-IP) analyses revealed that TRIM37 interacted with DUSP6, and that TRIM37 overexpression enhanced DUSP6 ubiquitination in GC cells. In vivo experiments showed the inhibitory effect of TRIM37 knockdown on tumor growth. These findings suggested that TRIM37 may act as an oncogene in GC to regulate c-myc, cleaved caspase 3, and the ERK1/2 pathway.
Patients and Methods
Patients and Tissues
Ninety-five GC patients aged 40 to 60 years old, who were treated at Renji Hospital, School of Medicine, Shanghai Jiao Tong University (Shanghai, China) were enrolled in this study. After informed consent was obtained, 30 paired cancer and paracancer tissues from 30 GC patients were collected to detect TRIM37 expression. After 80 months, the median follow-up time, samples of 65 GC patients were subjected to Kaplan-Meier survival analysis and Log rank test. This study was performed in accordance with the rules of the Declaration of Helsinki and approved by the Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University (Shanghai, China).
Cell Culture
Five human GC cell lines AGS, HGC27, MKN28, MKN45, SNU719 and a gastric mucosa cell line, GES-1, were purchased from the Cell Bank of Chinese Academy of Science (Shanghai, China). The above cells were cultured with RPMI-1640 medium (SH30809.01B, HyClone), containing 10% fetal bovine serum (FBS; 16,000–044, GIBCO) and 1% penicillin and streptomycin (P1400-100, Solarbio) at 37°C in a 5% CO2 incubator.
Construction of Lentivirus
Targeting three different sites of TRIM37 (shown in Table 1), interference sequences were synthesized (T1300-100, Solarbio) and shRNA was constructed by double chain annealing. Subsequently, each of the shRNAs was inserted into the AgeI/EcoRI sites of the pLKO.1-puro vector to construct pLKO.1-puro-shTRIM37-1, −2, and −3. For TRIM37 overexpression plasmids, the full length 2895 bp coding DNA sequence (CDS) region of TRIM37 (BC036012.1) was synthesized by Genewiz Company (Shanghai, China). Then, TRIM37 CDS was inserted into a pLVX-puro vector at the EcoR I/BamH I sites. After sequence confirmation, (Invitrogen, Thermo Fisher Scientific, Inc., USA), pLKO.1-puro-shTRIM37 (1000 ng) or pLVX-puro-TRIM37 (1000 ng) was co-transfected with viral packaging plasmids, psPAX2 (100 ng) and pMD2G (900 ng) (Addgene, Inc., Cambridge, MA, USA) using Lipofectamine 2000TM into 293T cells. After 48 hours of transfection, viral particles in supernatants were obtained by ultracentrifugation.
 |
Table 1 TRIM37 Interference Sequences |
Cell Proliferation Assay
GC cells (HGC27, MKN45 and AGS) were grown to the logarithmic phase, trypsinized (T1300-100, Solarbio) and counted under a microscope to prepare a cell suspension with 3 × 104 cells/mL. Then, 100 µL of cell suspension was taken to seed in 96-well plates and cultured overnight. After treatment at 0, 24, 48 and 72 hours, Cell Count Kit-8 (CCK-8; CP002, SAB) and serum-free medium were mixed at a volume ratio of 1:10 and then 100 µL was added to each group and incubated for 1 hour at 37°C. The the absorbance value (OD) at 450 nm was evaluated on a microplate reader.
Cell Cycle Detection
GC cells (HGC27, MKN45 and AGS) were grown to the logarithmic phase, trypsinized (T1300-100, Solarbio) and then seeded at 3 × 105 cells/well into 6-well plates for overnight culture. After treatment for 48 hours, the cells were collected to carry out cell cycle detection. In brief, cells were trypsinized and centrifuged for 5 min at 1000 g. Cells were then washed with pre-cooled PBS and then resuspended in 300 µL FBS-containing PBS (16,000–044, GIBCO), followed by 24 hours of immobilization in 700 μL of −20°C pre-cooled absolute ethanol at 4°C. After removing RNA with a 1 mg/mL RNase A solution (100 µL; R8020-25, Solarbio) at 37°C in the dark for 30 min, cells were stained in 400 µL Propidium Iodide (PI, 50 µg/mL; C001-200, 7Seabiotech, Shanghai, China) in the dark for 10 min. Within 24 hours, the DNA at each cell cycle was detected by a flow cytometer (Accuri C6, BD Biosciences) at a wavelength of 488 nm of the excitation wavelength. Cell cycle analysis was then performed using FLOWJO software.
Cell Apoptosis Assay
GC cells (HGC27, MKN45 and AGS) were grown to the logarithmic phase, trypsinized (T1300-100, Solarbio) and cultured overnight in 6-well plates with 3 × 105 cells/well. After treatment for 48 hours, the cells were trypsinized to collect in a centrifuge tube for apoptosis detection (Annexin V-FITC Apoptosis Detection Kit, C1063, Beyotime). About 1 × 105 resuspended cells were incubated with 195 μL of Annexin V-FITC binding solution, followed by 15 min incubation with 5 µL of Annexin V-FITC at 4°C in the dark. Thereafter, at 4°C in the dark, cells were incubated for 5 min in 5 µL PI staining solution. A tube without Annexin V-FITC and PI was used as a negative control. Subsequently, flow cytometry was used to detect cell apoptosis (FCM; Accuri C6, BD Biosciences).
Real-Time Polymerase Chain Reaction (RT-PCR) Assay
After treatment or without treatment, GC tissues or cells were collected to isolate the total RNA by Trizol Reagent (1596–026, Invitrogen). After quantification and confirmation of RNA integrity and purity, about 1 µg of RNA was reverse transcribed into complementary DNA (cDNA) by a Reverse Transcription Kit (#K1622, Fermentas, USA). Using a SYBRGreen PCR Kit, RT-PCR reactions using the cDNA as a template, were performed on the 7300 SDS Software of a Real-time detector (ABI-7300, Applied Biosystems, USA) with the following program: 95°C, 10min; (95°C, 15 sec; 60°C, 45 sec) × 40. After that, the expression of TRIM37 mRNA, normalized to GAPDH, was analyzed using the 2−ΔΔCq method.22 The primers are as listed: TRIM37, Forward: 5ʹ-TGGACTTACTCGCAAATG-3ʹ, Reverse: 5ʹ-ATCTGGTGGTGACAAATC-3ʹ; GAPDH, Forward: 5ʹ-AATCCCATCACCATCTTC-3ʹ, Reverse: 5ʹ-AGGCTGTTGTCATACTTC-3ʹ.
Western Blotting Analysis
After treatment or without treatment, GC cells were lysed in RIPA Lysis Buffer (P0013B, Beyotime), containing protease and phosphatase inhibitor, and then centrifuged at 12,000 g for 15 min and supernatant-containing protein was collected. Proteins were quantified with a BCA Kit (PICPI23223, Thermo Fisher Scientific, Inc., USA) prior to separation by electrophoresis using a 15% PAGE-SDS gel, and then transferred onto polyvinylidene fluoride (PVDF) membranes using a semi-dry transfer. Membranes were then blocked for 1 hour at room temperature in skim milk (BD Biosciences, Franklin Lakes, NJ, USA), followed by incubation at 4°C with primary antibodies against TRIM37 (1:1000; Ab95997, Abcam), c-myc (1:1000; Ab32072, Abcam), cleaved caspase 3 (1:500; Ab2302, Abcam), ERK1/2 (1:1000, #9102, Cell Signaling Technology [CST]), p-ERK1/2 (1:1000, #9101, CST) and GAPDH (1:2000, #5174, CST) overnight with gentle shaking. Thereafter, membranes were washed with TBST 3 times and incubated at 37°C for 1 hour with HRP-conjugated goat anti-rabbit secondary (1:1000; A0208, Beyotime, Haimen, China). After washing, a chemiluminescent reagent (WBKLS0100, EMD Millipore) was used to develop the membranes. Membranes were then exposed on an ECL imaging system (Tanon-5200, Tanon, Shanghai, China). Protein levels of TRIM37, c-myc, cleaved caspase 3, ERK1/2 and p-ERK1/2 were normalized to GAPDH and were then analyzed using ImageJ of version 1.47 (Bethesda, MD, USA).
Hematoxylin-Eosin (HE) Staining
About 0.3 cm of tumor tissues of a nude mouse were collected, fixed, embedded and cut into 4 µm slices by a paraffin slicer. After that, the slices were stained hematoxylin and eosin (BASO). In brief, the slides were baked for 30 min in an oven at 65°C and then dewaxed in xylene I and II (10,023,418, Sinopharm) for 15 min in turns. Rehydration was carried out in 100%, 95%, 85% and 75% ethyl alcohol (10,092,680, Sinopharm) for 5 min in turns followed by a rinse with tap water for 10 min. Sections were then stained with hematoxylin (714,094, BASO) for 5 min, followed by 2 secs of color separation in amino water (10,002,118, Sinopharm) and 15 min rinsing with tap water. Sections were then dehydrated in 70% and 90% ethyl alcohol for 10 min in turns and then stained with eosin (BA4099, BASO) for 2 min, followed by dehydration with absolute ethanol. 3 sections were hyalinized in xylene for 3 min twice, sealed with neutral gum (G8590, Solarbio), and placed in a 65°C oven for 15 min. Subsequently, sections were imaged using a microscopic image analysis system (DS-Ri2, NIKON), and the relevant parts of the samples were collected and analyzed.
TUNEL Fluorescence
About 0.3 cm of tumor tissues of a nude mouse were collected, fixed, embedded and cut into 4 µm slices by a paraffin slicer. Later, sections were used to detect cell apoptosis of tumor tissues by a TUNEL Apoptosis Detection Kit (FY600017-20T, Fuyuanbio) following the manufacturer’s instructions. Finally, the slides were imaged on a fluorescence microscope.
Immunohistochemical (IHC) Detection
About 0.3 cm of cancer and paracancer tissues of GC patients were collected, fixed, embedded and cut into 4 µm slices by a paraffin slicer. After being baked for 30 min in an oven at 65°C, sections were dewaxed in xylene I and xylene II (Shanghai Sinopharm) for 15 min in order, and then rehydrated sequentially in gradient concentrations of 100%, 95%, 85%, and 75% ethanol for 5 min, and then rinsed for 10 min with tap water. After that, antigen retrieval was carried out for 15 min in 0.01 M sodium citrate buffer (pH6.0), and sections were then blocked in a wet-box with 0.3% H2O2. Following incubation with Rb-TRIM37 antibody for 1 hour at room temperature, sections were incubated with HRP-conjugated secondary antibody. Sections were then subjected to DAB staining, 3 min staining of hematoxylin and alcohol differentiation with 1% hydrochloric acid. Following a 10 min rinse with tap water, sections were hyalinized in xylene for 3 min × 2 times, sealed with neutral gum (G8590, Solarbio), and placed in a 65°C oven for 15 min. Subsequently, pictures were taken using a microscopic image analysis system (DS-Ri2, NIKON), and the relevant parts of the samples were collected and analyzed. According to the positive areas, the specimens were categorized into two groups: TRIM37 low expression: < 25% of the tumor cells showed positive TRIM37; TRIM37 high expression: > 25% of the tumor cells showed positive TRIM37. For evaluation criteria of IHC results, refer to the report of Chen et al.23
Co-Immunoprecipitation (Co-IP)
Briefly, whole-cell extracts were isolated after infection with vector or oeTRIM37 lentivirus. Then, all samples were incubated with the appropriate antibodies plus Protein A/G beads (Santa Cruz Biotechnology, USA) overnight. Beads were washed five times and separated by SDS-PAGE. Western blot was performed as indicated above.
Ubiquitination Assay
AGS cells that were infected with vector or oeTRIM37 lentivirus were lysed by sonication in 1% SDS-containing radio immunoprecipitation assay (RIPA) buffer on ice. Next, lysates were treated with Protein A/G PLUS-Agarose (sc-2003, Santa Cruz Biotechnology, USA) for 1 h. Then, the samples were incubated with IgG (sc-2027, Santa Cruz Biotechnology, USA) overnight at 4°C. After centrifugation for 5 min at 3000 rpm at 4°C, the nuclear pellet was gathered and washed four times with Protein A/G Plus-Agarose beads. The purified proteins were run on a 4–20% gradient SDS-PAGE. Anti-DUSP6 antibody (ab76310, Abcam, UK) and anti-Ubiquitin antibody (ab7780, Abcam, UK) were used for immunoblotting.
In vivo Experiments
The BALB/c nude mice were divided into two groups: HGC27-shTRIM37 and HGC27-shNC (normal control). 0.1 mL of PBS suspended with 1×106 cells was subcutaneously injected into nude mice (BALB/c, male, 4 weeks old), respectively. Tumor volume was measured every third day. The mice were euthanized 4 weeks later. The tumor growth and apoptosis was detected by HE staining and TUNEL fluorescence. All animal protocols were carried out following the Guidelines for Care and Use of Laboratory Animals (Ministry of Science and Technology of China) and authorized by the Institutional Animal Ethics Care and Use Committee of the Renji Hospital, School of Medicine, Shanghai Jiao Tong University.
Statistical Analysis
Statistical analysis in this research was conducted on the GraphPad prism 7.0 software (GraphPad Software, USA). With three repeated experiments, all graphed results are presented as mean ± SD. Paired Student’s t-test was used to determine the significance between two groups, while multiple groups were determined by One-way analysis of variance (ANOVA) with Tukey’s post hoc test. A value of p < 0.05 was considered statistically significant.
Results
TRIM37 Was Highly Expressed in Tumor Tissues of GC Patients and GC Cell Lines
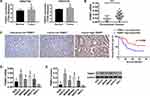
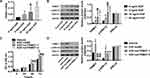
Analysis of normal and tumor samples from TCGA database showed that expression of TRIM37 in GC tumors was much higher than that in normal tissues (Figure 1A). In our study, thirty paired cancer and paracancer tissues from GC patients were collected to analyze the expression of TRIM37. As shown in Figure 1B, compared to paracancer tissues, the mRNA expression of TRIM37 in cancer tissues of GC patients was significantly increased. IHC staining of 65 GC patients also showed high protein expression of TRIM37 in cancer tissues. In GC tissues, the staining intensity of TRIM37 is significantly higher than that of the corresponding adjacent tissue, and the expression of TRIM37 is found in both cytoplasm and nucleus. After 80 months, 37 of 65 patients died of GC. Kaplan-Meier survival analysis and Log rank test demonstrated that TRIM37 expression was significantly correlated with overall survival, and patients with high expression of TRIM37 had a poor prognosis (Figure 1C). Relationship between TRIM37 expression and clinicopathological features of gastric cancer was shown in Table 2. Consistent with the above observation, we found significantly higher expression of TRIM37 in GC cell lines (AGS, HGC27, MKN28, MKN45 and SNU719) compared with the gastric mucosa cell line, GES-1 (Figure 1D and E). Furthermore, compared with other cell lines, TRIM37 is relatively high in HGC27 and MKN45, and relatively low in AGS. These findings suggested that TRIM37 may act as an oncogene in GC.
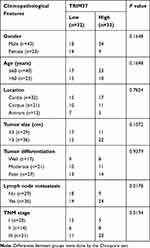 |
Table 2 Relationship Between TRIM37 Expression and Clinicopathological Features of Gastric Cancer |
Knockdown and Overexpression of TRIM37 in GC Cells by Infection with Lentivirus
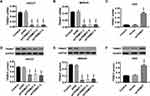
In vitro, two GC cell lines, HGC27 and MKN45, were infected with shTRIM37 lentiviruses (shTRIM37-1, −2 and −3), while AGS cells were infected oeTRIM37 lentivirus. After RT-PCR and Western blotting analysis, the results showed that all three shTRIM37 lentiviruses significantly down-regulated the expression of TRIM37 mRNA in HGC27 (Figure 2A and D) and MKN45 (Figure 2B and E) cells, while oeTRIM37 lentivirus significantly up-regulated TRIM37 expression in AGS cells (Figure 2C and F). In addition, compared to shTRIM37-3, lentiviruses of shTRIM37-1 and −2 had a more profound effect. Therefore, due to the effectiveness of the knockdown or overexpression, lentivirus of shTRIM37-1, −2 and oeTRIM37 were used for follow-up experiments.
Knockdown of TRIM37 in GC Cells Suppressed Cell Proliferation, Cell Cycle Progression and Promoted Apoptosis
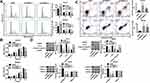
To understand the function of TRIM37 in GC cells, proliferation, cell cycle and apoptosis were evaluated after TRIM37 knockdown. The results in Figure 3A showed that knockdown of TRIM37 significantly prevented cell cycle progression by arresting cells in the G1 phase, thereby reducing cell entry into the S/G2 phase. Furthermore, in TRIM37-silenced HGC27 and MKN45 cells, cell proliferation was significantly decreased (Figure 3B), whereas apoptosis was increased (Figure 3C), concurrent with increased cleaved caspase 3 and decreased c-myc, as well as decreased ERK1/2 phosphorylation, while ERK1/2 remained unchanged (Figure 3D). These results indicated that knockdown of TRIM37 could suppress cell proliferation in GC cancer, likely by blocking cell cycle progression and promoting apoptosis.
TRIM37 Regulation of Proliferation, Cell Cycle Progression and Apoptosis in GC Cells May Be Mediated by ERK1/2 Signaling Pathway
We also investigated the underlying mechanism of TRIM37 in regulating GC cells. As shown in Figure 4A, overexpression of TRIM37 in AGS cells significantly promoted the progression of the cell cycle from the G1-phase to the S-phase. Concurrently, cell proliferation in AGS was significantly increased by TRIM37 overexpression (Figure 4B), while apoptosis was decreased (Figure 4C). This was accompanied by decreased cleaved caspase 3 and increased c-myc and phosphorylated ERK1/2, while ERK1/2 remained unchanged (Figure 4D). In addition, the induction of TRIM37 overexpression was potently attenuated by the ERK1/2 inhibitor, U0126. Taken together, we speculated that overexpression of TRIM37 may contribute to GC progression, possibly through the activation of the ERK1/2 signaling pathway.
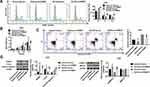
Knockdown of TRIM37 Potently Attenuated EGF-Induced Cell Proliferation in GC Cells
A series of concentrations of EGF recombinant proteins (ERK1/2 agonist) were used to treat AGS cells. As shown in Figure 5A and B, EGF recombinant proteins significantly increased TRIM37 expression and ERK1/2 phosphorylation in a dose-dependent manner. Furthermore, EGF-induced (10 ng/mL) GC cell proliferation was potently attenuated by TRIM37 knockdown (Figure 5C). Likewise, EGF-induced TRIM37 expression and ERK1/2 phosphorylation was significantly decreased by TRIM37 knockdown, while ERK1/2 expression was unchanged (Figure 5D). These data further prove that the ERK1/2 signaling pathway is involved in the regulation of GC cells by TRIM37.
TRIM37 Interacted with DUSP6 and Enhanced Its Ubiquitination in GC Cells
Next, we examined the mRNA and protein levels of DUSP6 in GC cells infected with oeTRIM37. Interestingly, the relative DUSP6 mRNA showed no significant change (Figure 6A), however, the DUSP6 protein was significantly down-regulated in oeTRIM37-infected GC cells (Figure 6B). This demonstrated that overexpression of TRIM37 did not affect DUSP6 transcription, but instead, suppressed its translation in GC cells. In addition, Co-IP assays showed the interaction between TRIM37 and DUSP6 in GC cells (Figure 6C). TRIM37 overexpression significantly enhanced DUSP6 ubiquitination in GC cells, suggesting that TRIM37 might suppress DUSP6 translation by enhancing its ubiquitination in GC cells (Figure 6D).
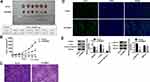
Knockdown of TRIM37 in Nude Mice Significantly Inhibited Tumor Growth
In vivo experiments were also performed on nude mice. We found that knockdown of TRIM37 significantly inhibited tumor weight (Figure 7A) and volume (Figure 7B) of nude mice. HE (Figure 7C) and TUNEL (Figure 7D) staining of tumor tissues showed a significant increased in cell apoptosis. Moreover, Western blotting showed that in TRIM37-silenced tumor tissues, protein levels of cleaved caspase 3 were significantly increased, and c-myc and phosphorylated ERK1/2 were decreased, while ERK1/2 was unchanged (Figure 7E). These demonstrated that knockdown of TRIM37 in nude mice significantly inhibited tumor growth, which further indicated the inhibitory effect of TRIM37 knockdown and the ERK 1/2 signaling pathway in GC progression.
Discussion
Although the incidence and mortality of gastric cancer have been reduced in the US and elsewhere, it is still a major public health concern. In recent years, increased research showed the involvement of TRIM proteins in a variety of human cancers, including gastric cancer. For example, expression of TRIM29, TRIM44 and TRIM26 are associated with the poor prognosis and overall survival of cancer patients.24–26 Studies have found that TRIM59 and TRIM31 are up-regulated in GC,27,28 and TRIM29 acts as an oncogene in GC and is a new marker for lymph node metastasis.29,30 In addition, down-regulation of TRIM25 can suppress GC cell migration and invasion.31 In this study, we found a significant increase in the expression of TRIM37 in tumor tissues of GC patients and GC cell lines, and patients with high expression of TRIM37 had a poor prognosis. Knockdown of TRIM37 significantly inhibited cell proliferation, blocked cell cycle progression at the G1-phase and promoted apoptosis, whereas TRIM37 overexpression had the opposite effect. Our results are consistent with previous reports that knockdown of TRIM37 can suppress cell proliferation and tumor growth in breast cancer.9 These findings suggested that TRIM37 may act as an oncogene in GC, and knockdown of TRIM37 may be considered for GC treatment.
Furthermore, we explored the potential mechanisms of TRIM37 in regulating GC cell proliferation, cell cycle and apoptosis. We found that expression of cleaved caspase 3 was increased, while c-myc was decreased in TRIM37-silenced GC cells. C-myc is a pro-oncogene that can regulate cell proliferation, promote cell division and participate in cell apoptosis, and is associated with the development of various tumors.32,33 Cleaved caspase 3 is a cysteine protease regulating apoptosis or programmed cell death.34 Thus, we speculated that TRIM37 regulation of cell proliferation, cell cycle progression and apoptosis may be mediated by the modulation of cleaved caspase 3 and c-myc. Previous studies have reported that β-catenin signaling is frequently activated in GC, which is associated with the survival of GC patients.35,36 The TCF-β signaling pathway is involved in the regulation of TRIM25 in GC cell migration and invasion.31 In our study, knockdown of TRIM37 significantly reduced ERK1/2 phosphorylation, while TRIM37 overexpression had the opposite effect. Furthermore, TRIM37-induced phosphorylation of ERK1/2 and c-myc was potently decreased by an ERK1/2 inhibitor, U0126, while cleaved caspase 3 was increased. TRIM37 knockdown potently reduced cell proliferation, as well as the phosphorylation of ERK1/2 which was induced by an ERK1/2 agonist, EGF recombinant protein. Moreover, we found that TRIM37 overexpression did not affect DUSP6 transcription, but suppressed its translation in GC cells. Further analyses demonstrated the interaction between TRIM37 and DUSP6, and TRIM37 overexpression enhanced DUSP6 ubiquitination in GC cells. Therefore, TRIM37 overexpression might suppress DUSP6 protein levels by enhancing its ubiquitination and degradation in GC cells. In vivo experiments in nude mice further showed the inhibitory effect of TRIM37 knockdown on tumor growth, accompanied with increased cleaved caspase 3 and decreased c-myc and phosphorylated ERK1/2. Taken together, we inferred that TRIM37 regulated GC cell proliferation, cell cycle and apoptosis likely by activating the ERK1/2 pathway via the modulation of cleaved caspase 3 and c-myc, which is in agreement with reports that the ERK1/2 signaling pathway participates in GC.37,38 However, the lack of specific mechanisms of TRIM37-ERK1/2 in regulation of caspase 3 and c-myc in GC is one of the limitations of this study. If possible, further experiments exploring this aspect will be carried out in the future. In summary, this research demonstrates that TRIM37 likely functions as an oncogene in GC. Knockdown of TRIM37 can significantly suppress GC cell proliferation, arrest cell cycle progression at the G1-phase and promote apoptosis, likely mediated by the activation of the ERK1/2 signaling pathway via the modulation of cleaved caspase 3 and c-myc. Thus, targeting TRIM37 may serve as a potential novel therapy for GC treatment.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Funding
This study is supported by grant from Shanghai Municipal Health Commission (201940433).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi:10.3322/caac.21208
2. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi:10.3322/canjclin.55.2.74
3. Herszenyi L, Tulassay Z. Epidemiology of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci. 2010;14:249–258.
4. Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388(10060):2606. doi:10.1016/S0140-6736(16)32226-7
5. Roder DM. The epidemiology of gastric cancer. Gastric Cancer. 2002;5(S1):5–11. doi:10.1007/s10120-002-0203-6
6. Martin RC, Jaques DP
7. Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem Sci. 2012;37:58–65. doi:10.1016/j.tibs.2011.11.001
8. Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat. 2003;78:313–322. doi:10.1023/A:1023081624133
9. Bhatnagar S, Gazin C, Chamberlain L, et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116.
10. Jiang J, Tian S, Yu C, Chen M, Sun C. TRIM37 promoted the growth and migration of the pancreatic cancer cells. Tumor Biol. 2016;37:2629–2634. doi:10.1007/s13277-015-4078-7
11. Tang S-L, Gao Y-L, Wen-zhong H. Knockdown of TRIM37 suppresses the proliferation, migration and invasion of glioma cells through the inactivation of PI3K/Akt signaling pathway. Biochem Pharmacol. 2018;99:59–64. doi:10.1016/j.biopha.2018.01.054
12. Jiang J, Yu C, Chen M, Tian S, Sun C. Over-expression of TRIM37 promotes cell migration and metastasis in hepatocellular carcinoma by activating Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2015;464:1120–1127. doi:10.1016/j.bbrc.2015.07.089
13. Zhao P, Guan H-T, Dai Z-J, Ma Y-G, Liu -X-X, Wang X-J. Knockdown of tripartite motif-containing protein 37 (TRIM37) inhibits the proliferation and tumorigenesis in colorectal cancer cells. Oncol Res. 2017;25(1):115–122. doi:10.3727/096504016X14732772150181
14. Hu C-E, Gan J. TRIM37 promotes epithelial-mesenchymal transition in colorectal cancer. Mol Med Rep. 2017;15(3):1057–1062. doi:10.3892/mmr.2017.6125
15. Arkell RS, Dickinson RJ, Squires M, Hayat S, Keyse SM, Cook SJ. DUSP6/MKP-3 inactivates ERK1/2 but fails to bind and inactivate ERK5. Cell Signal. 2008;20(5):836–843. doi:10.1016/j.cellsig.2007.12.014
16. Moncho-Amor V, Pintado-Berninches L, Ibañez de Cáceres I, et al. Role of Dusp6 phosphatase as a tumor suppressor in non-small cell lung cancer. Int J Mol Sci. 2019;20(8):2036. doi:10.3390/ijms20082036
17. Okudela K, Yazawa T, Woo T, et al. Down-regulation of DUSP6 expression in lung cancer: its mechanism and potential role in carcinogenesis. Am J Pathol. 2009;175(2):867–881. doi:10.2353/ajpath.2009.080489
18. Wang L, Yuqiu Z, Wenli F, et al. Dual specificity phosphatase 6 protects neural stem cells from β-amyloid-induced cytotoxicity through ERK1/2 inactivation. Biomolecules. 2018;8(4):181.
19. Ma J, Yu X, Guo L, Lu S. DUSP6, a tumor suppressor, is involved in differentiation and apoptosis in esophageal squamous cell carcinoma. Oncol Lett. 2013;6:1624–1630.
20. Xia H, Zhenghao T, Siyuan M, et al. Tripartite motif-containing protein 7 regulates hepatocellular carcinoma cell proliferation via the DUSP6/p38 pathway. Biochem Biophys Res Commun. 2019;511(4):889–895.
21. Di K, Linskey ME, Bota DA. TRIM11 is overexpressed in high-grade gliomas and promotes proliferation, invasion, migration and glial tumor growth. Oncogene. 2013;32(42):5038–5047. doi:10.1038/onc.2012.531
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25:402–408. doi:10.1006/meth.2001.1262
23. Chen D, You X, Pan Y, Liu Q, Cao G. TRIM37 promotes cell invasion and metastasis by regulating SIP1-mediated epithelial-mesenchymal transition in gastric cancer. Onco Targets Ther. 2018;11:8803
24. Palmbos PL, Wang L, Yang H, et al. ATDC/TRIM29 drives invasive bladder cancer formation through miRNA-mediated and epigenetic mechanisms. Cancer Res. 2015;75(23):5155–5166. doi:10.1158/0008-5472.CAN-15-0603
25. Zhu X, Wu Y, Miao X, et al. High expression of TRIM44 is associated with enhanced cell proliferation, migration, invasion, and resistance to doxorubicin in hepatocellular carcinoma. Tumor Biol. 2016;37(11):14615–14628. doi:10.1007/s13277-016-5316-3
26. Wang Y, He D, Yang L, et al. TRIM26 functions as a novel tumor suppressor of hepatocellular carcinoma and its downregulation contributes to worse prognosis. Biochem Biophys Res Commun. 2015;463(3):458–465. doi:10.1016/j.bbrc.2015.05.117
27. Zhou Z, Ji Z, Wang Y, et al. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology. 2014;147(5):1043–1054. doi:10.1053/j.gastro.2014.07.021
28. Sugiura T. The cellular level of TRIM31, an RBCC protein overexpressed in gastric cancer, is regulated by multiple mechanisms including the ubiquitin—proteasome system. Cell Biol Int. 2011;35:657–661. doi:10.1042/CBI20100772
29. Qiu F, Xiong J-P, Deng J, Xiang X-J. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int J Clin Exp Pathol. 2015;8:5053.
30. Kosaka Y, Inoue H, Ohmachi T, et al. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14:2543–2549. doi:10.1245/s10434-007-9461-1
31. Zhu Z, Wang Y, Zhang C, et al. TRIM25 blockade by RNA interference inhibited migration and invasion of gastric cancer cells through TGF-β signaling. Sci Rep. 2016;6:19070.
32. Evan GI, Wyllie AH, Gilbert CS, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi:10.1016/0092-8674(92)90123-T
33. Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi:10.1128/MCB.19.1.1
34. Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi:10.1042/bj3260001
35. Clements WM, Wang J, Sarnaik A, et al. β-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002;62:3503–3506.
36. Zhou Y-N, Xu C-P, Han B, et al. Expression of E-cadherin and β-catenin in gastric carcinoma and its correlation with the clinicopathological features and patient survival. World J Gastroenterol. 2002;8:987. doi:10.3748/wjg.v8.i6.987
37. Zhou Q, Wang X, Yu Z, et al. Transducin (β)-like 1 X-linked receptor 1 promotes gastric cancer progression via the ERK1/2 pathway. Oncogene. 2017;36:1873. doi:10.1038/onc.2016.352
38. Ertao Z, Jianhui C, Chuangqi C, et al. Autocrine sonic hedgehog signaling promotes gastric cancer proliferation through induction of phospholipase Cγ1 and the ERK1/2 pathway. J Exp Clin Cancer Res. 2016;35:63. doi:10.1186/s13046-016-0336-9
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.