Back to Journals » OncoTargets and Therapy » Volume 11
Knockdown of RNF6 inhibits gastric cancer cell growth by suppressing STAT3 signaling
Authors Huang Z, Cai Y, Yang C, Chen Z, Sun H, Xu Y, Chen W, Xu D, Tian W, Wang H
Received 21 May 2018
Accepted for publication 20 August 2018
Published 5 October 2018 Volume 2018:11 Pages 6579—6587
DOI https://doi.org/10.2147/OTT.S174846
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Arseniy Yuzhalin
Ziming Huang,1 Yong Cai,1 Chenchen Yang,1 Zhen Chen,1 Hong Sun,1 Yingying Xu,2 Wei Chen,3 Dafu Xu,4 Wenze Tian,4 Haixiao Wang5
1Department of Emergency Surgery, The Affiliated Huaian No 1 People’s Hospital of Nanjing Medical University, Huai’an 223300, Jiangsu, China; 2Department of Radiotherapy, The Affiliated Huaian No 1 People’s Hospital of Nanjing Medical University, Huai’an 223300, Jiangsu, China; 3Department of Respiratory Care, The Affiliated Huaian No 1 People’s Hospital of Nanjing Medical University, Huai’an 223300, Jiangsu, China; 4Department of Cardiothoracic Surgery, The Affiliated Huaian No 1 People’s Hospital of Nanjing Medical University, Huai’an 223300, Jiangsu, China; 5Department of General Surgery, The Affiliated Huaian No 1 People’s Hospital of Nanjing Medical University, Huai’an 223300, Jiangsu, China
Background and objective: RNF6, an E3 ligase, has been reported to play an important role in the tumorigenesis in several tissues, but its role in gastric cancer is still unknown. In this study, we aimed to investigate the biological function and molecular mechanisms of RNF6 in gastric cancer.
Materials and methods: The expression levels of RNF6 were detected by quantitative real-time PCR (qRT-PCR) and immunoblotting in gastric cancer tissues and cell lines. Cell Counting Kit-8 assay was performed to evaluate cell proliferation. Cell apoptosis was analyzed by flow cytometer and immunoblotting. Luciferase assay, immunoblotting and qRT-PCR were performed to explore the activation of STAT3. Immunoprecipitation was performed to evaluate the ubiquitination of SHP-1.
Results: In this study, RNF6 was found to be upregulated in both primary tissues and cell lines of gastric cancer. Knockdown or overexpression of RNF6 inhibited or promoted cell growth of gastric cancer cells. Knockdown of RNF6 also induced the cleavage of PARP and promoted cell apoptosis in gastric cancer cells. In addition, knockdown of RNF6 also increased the cytotoxicity of doxorubicin against gastric cancer. Moreover, knockdown of RNF6 inhibited STAT3-derived luciferase activity and downregulated the phosphorylation of STAT3, but upregulated the protein level of SHP-1. Knockdown of RNF6 downregulated the expression of MCL1 and XIAP, which are target genes of STAT3. Further studies showed that RNF6 regulated the stability of SHP-1 by inducing its polyubiquitination.
Conclusion: These results demonstrated that RNF6 was highly expressed in gastric cancer and regulated the growth of gastric cancer cells by affecting SHP-1/STAT3 signaling, which suggested that RNF6 could be a novel target for gastric cancer therapy.
Keywords: RNF6, cell growth, gastric cancer, STAT3, SHP-1
Introduction
Gastric cancer is the most common gastrointestinal malignancy and a leading cause of cancer-related deaths worldwide.1 There are about 700,000 confirmed mortalities annually worldwide.2 Although therapy regimens for gastric cancer include surgery, radiation, chemotherapy or a combination, it is still difficult to treat gastric cancer in clinic where it is often found late.3 Therefore, there is an urgent demand to identify new targets and drugs to improve systemic therapy for gastric cancer patients.
RNF6 belongs to the largest E3 ligase family and plays an important role in the tumorigenesis in several tissues.4 At first, RNF6 was considered a tumor suppressor because of its mutations and its location on chromosome 13q12 in human esophageal squamous cell carcinoma.5 But recent studies have indicated that RNF6 is more likely an oncogene.
Recent studies showed that RNF6 was upregulated in colorectal cancer and promoted colorectal tumorigenesis by activating Wnt/β-catenin pathway or JAK/STAT3 pathway.6,7 In leukemia, RNF6, as a direct target of the transcription factor PBX1, was overexpressed and induced leukemia cell growth.8 RNF6 was also upregulated in breast cancer and predicted a poor prognosis of breast cancer patients.9 RNF6 promoted breast cancer cell growth by increasing the stability of estrogen receptor alpha (ERα), thus targeting the RNF6/ERα/Bcl-xL pathway may be a promising strategy for breast cancer treatment.9 RNF6 was also found highly expressed in prostate cancer and promoted the transcriptional activity of androgen receptor (AR) by mediating its atypical polyubiquitination at Lys-6 and Lys-27.10 However, the studies on RNF6 are very limited, and the biological function of RNF6 is still unknown in most tumors, including gastric cancer.
In this study, we evaluated the function of RNF6 in gastric cancer cells and found that RNF6 was upregulated in gastric cancer cells and contributed to gastric cancer cell growth. Moreover, knockdown of RNF6 suppressed the phosphorylation of STAT3 in gastric cancer cells, but upregulated the protein level of SHP-1, a negative regulator of STAT3. In addition, decreased RNF6 enhanced the cytotoxicity of doxorubicin (DOX) against gastric cancer cells.
Materials and methods
Cells, tissues and chemicals
Gastric cancer cell lines such as AGS, HGC-27, MGC80-3, NCI-N87 and SNU-1 and a human normal gastric mucosal cell line were purchased from Shanghai Cell Institute of Chinese Academy of Sciences (Shanghai, China). HEK293T cell line was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All gastric cancer cell lines, human normal gastric mucosal cell line and HEK293T were maintained in DMEM with 10% FBS, 100 U/mL of streptomycin and 100 μg/mL of penicillin. Mycoplasma tests of the cell lines used in this study have been carried out before starting the experiments. DOX was purchased from Sigma-Aldrich Co. (St Louis, MO, USA).
Quantitative real-time PCR (qRT-PCR)
The qRT-PCR was performed as previously described.11 First, total RNA was extracted by Trizol reagent according to the manufacturer’s instructions (Takara Bio Group, Kusatsu, Japan). Then, cDNA was synthesized from equal quantities of total RNA by PrimeScript™ RT reagent Kit (Takara Bio Group). Subsequently, to determine the mRNA levels of RNF6, MCL1 and XIAP, qRT-PCR was performed by using SYBR Green qPCR Master Mix (Takara Bio Group) on a Roche LightCycler® 480II real-time PCR system (Hoffman-La Roche Ltd., Basel, Switzerland). The primers used were as follows: RNF6, forward 5′-AGAAGATGGCAGCAAGAGCG-3′ and reverse 5′-TCAAGTCAGGCTGAGATGCTAGT-3′; MCL1, forward 5′-GCGACGGCGTAACAAACT-3′ and reverse 5′-ATTCCTGATGCCACCTTCTAG-3′; XIAP, forward 5′-TGGCAGATTATGAAGCAC-3′ and reverse 5′-CTCCTCCACAGTGAAAGC-3′; SHP-1, forward 5′-GAACGCTAAGACCTACATCG-3′ and reverse 5′-AGTATGGGACGCATTTGTT-3′; GAPDH, forward 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse 5′-TGGTGAAGACGCCAGTGGA-3′.
Immunoblotting
Immunoblotting analysis was performed as described previously.12–14 Whole cell lysates were lysed, and equal amounts of total proteins were subjected to SDS-PAGE separation, followed by immunoblotting with specific antibodies. The primary antibody against RNF6 was purchased from Thermo Fisher Scientific (Waltham, MA, USA; dilution: 1:1,000). Anti-PARP, cyclin D1, p-STAT3, STAT3, SHP-1, MCL1, Bcl-2 and GAPDH antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA; dilutions: 1:1,000). Anti-Ub antibody was purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA; dilution: 1:500). Anti-mouse and anti-rabbit immunoglobulin G (IgG) horseradish peroxidase-conjugated antibodies were purchased from Beyotime Biotechnology, Inc. (Nantong, China; dilutions: 1:1,000).
Lentivirus construction and infection
The lentivirus-delivered shRNAs against RNF6 (shRNF6) were synthesized from Suzhou GenePharma Co., Ltd. (Suzhou, China). The target sequences of shRNF6#1 and shRNF6#2 were 5′-TCAGGCAATTACCTTGCAT-3′ and 5′-ATAACAGTTCCTCTTCGTA-3′. Indicative lentiviral particles were produced in HEK293T cells and infected with NCI-N87 and SNU-1 cells.
Plasmid construction and gene transfection
The human RNF6 gene was amplified by PCR and cloned into pcDNA3.1 vector with a Myc tag as previously described.8 The primers for RNF6 amplification were as follows: forward 5′-ATGAATCAGTCTAGATCGAGATCAG-3′; reverse 5′-TTACCCATTGTTTGCTATGTTAGACCC-3′. Empty vector or Myc-RNF6 plasmids were transiently transfected into NCI-N87 cells by Lipofectamine® 2000 (Thermo Fisher Scientific) according to the manufacturer’s instruction.
Cell growth and viability
Viable cells were analyzed by Cell Counting Kit-8 (CCK-8) assay according to the manufacturer’s instructions (Biotool, Houston, TX, USA) as described previously.11 NCI-N87 cells, infected with shRNF6#1, shRNF6#2 or shNC, were treated with 0.5 μM DOX or dimethyl sulfoxide (DMSO) for 24 hours, followed by CCK-8 assay.
Cell apoptosis analysis
NCI-N87 and SNU-1 cells were infected with indicated lentivirus for 3 days, and then cells were collected for staining with Annexin V–fluorescein isothiocyanate (FITC) and propidium iodide (PI) for 5 minutes in the dark. Then, stained cells were prepared for cell apoptosis analysis on a flow cytometer (FACSCalibur®; BD Biosciences, San Jose, CA, USA) as described previously.15
Luciferase assay
NCI-N87 and SNU-1 cells were transfected with STAT3-Luc by Lipofectamine 2000 for 24 hours, followed by infection with shRNF6#1, shRNF6#2 or shNC. Three days later, cells were lysed for luciferase assay by using Dual-Luciferase® Reporter Assay System according to the manufacturer’s instructions (Promega Corporation, Fitchburg, WI, USA) as described previously.16
Immunoprecipitation analysis
The whole cell lysates were prepared for immunoprecipitation as described previously.14 First, the whole cell lysates were incubated with normal IgG or SHP-1 antibody overnight at 4°C, followed by incubation with protein A/G Sepharose beads (Santa Cruz Biotechnology Inc.) for 4 hours. The co-precipitated proteins were identified by immunoblotting analysis against Ub and SHP-1.
Statistical analyses
Student’s t-test was used for significant analysis in the studies. All statistical tests were two sided, and statistical significance was defined as a P-value of <0.05.
Results
RNF6 is overexpressed in gastric cancer and regulates gastric cancer cell growth
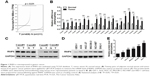
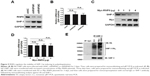
To evaluate the expression level of RNF6 in gastric cancer, The Cancer Genome Atlas database was first used for analysis, and, as shown in Figure 1A, RNF6 was obviously upregulated in tumor tissues of gastric cancer. Then, to confirm this result, 20 pairs of adjacent normal tissues and tumor tissues from gastric cancer patients were collected, and the qRT-PCR showed that the mRNA level of RNF6 was significantly higher in tumor tissues than the adjacent normal tissues (Figure 1B). The protein level of RNF6 was also upregulated in representative tumor tissues (Figure 1C). Moreover, RNF6 was upregulated in gastric cancer cell lines compared with normal gastric mucosal cell line (Figure 1D and E).
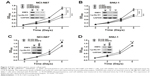
These results suggested that RNF6 was upregulated in both tumor tissues and gastric cancer cell lines; we then evaluated whether RNF6 regulated the cell growth of gastric cancer. As shown in Figure 2A and B, knockdown of RNF6 significantly inhibited the cell growth of both NCI-N87 and SNU-1 cells, along with cyclin D1 downregulation, which was involved in regulating cell cycle progression. In addition, the overexpression of RNF6 promoted the cell growth of both NCI-N87 and SNU-1 cells, along with cyclin D1 upregulation (Figure 2C and D). These results suggested that RNF6 was functional in gastric cancer and regulated the growth of gastric cancer cells.
Knockdown of RNF6 induces cell apoptosis and increases the cytotoxicity of DOX in gastric cancer
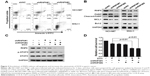
To find out whether cell apoptosis was also triggered by silenced RNF6, flow cytometry was performed by using Annexin V and propidium iodide staining. As shown in Figure 3A, compared with cells infected with shNC-derived lentivirus, cells infected with shRNF6#1 or shRNF6#2 harbored a significant increase in Annexin V+ in both NCI-N87 and SNU-1 cells. Immunoblotting analysis also revealed that knockdown of RNF6 induced the cleavage of PARP and inhibited the expression levels of anti-apoptotic protein MCL1 in both NCI-N87 and SNU-1 cells (Figure 3B), which further suggested that knockdown of RNF6 induced cell apoptosis in gastric cancer cells.
DOX is one of the most common chemotherapeutic drugs for many tumors, including gastric cancer, but frequent resistance was found in the process of tumor treatment in the clinic.17–19 A genetic approach was also used to explore the synergistic action of shRNF6 and DOX. Two shRNAs against RNF6 were used to knock down RNF6 in NCI-N87 cells (Figure 3C), and CCK-8 assay showed that the viability of NCI-N87 cells decreased only to ~80% when treated with DOX alone, but viability went down markedly to ~50% with synergistic action of shRNF6 and DOX (Figure 3D). Therefore, suppressing RNF6 enhanced the cytotoxic activity of DOX, and targeting RNF6 during DOX therapy had potential to improve outcomes.
Knockdown of RNF6 inhibits STAT3 signaling in gastric cancer
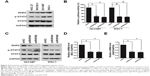
Recently, it was reported that RNF6 amplification activated STAT3 signaling in colorectal cancer,7 and we wanted to evaluate whether this occurred in gastric cancer. First, we collected four cell lines of gastric cancer, and we found that RNF6 and phospho-STAT3 were co-expressed, such that HGC-27 and MGC80-3 cells harbored lower levels of RNF6 and phospho-STAT3, but NCI-N87 and SNU-1 cells harbored higher levels (Figure 4A). Then, STAT3-derived luciferase assay was performed and results showed that knockdown of RNF6 suppressed the luciferase activity in both NCI-N87 and SNU-1 cells (Figure 4B). In addition, immunoblotting also revealed that knockdown of RNF6 downregulated the phosphorylation of STAT3 in both NCI-N87 and SNU-1 cells (Figure 4C). In addition, the downregulation of RNF6 by shRNA also downregulated the mRNA levels of STAT3 target genes MCL1 and XIAP (Figure 4D and E). These results suggested that RNF6 regulates STAT3 signaling in gastric cancer cells.
RNF6 regulates the stability of SHP-1 by inducing its polyubiquitination
SHP-1 is a non-transmembrane protein tyrosine phosphatase, and it serves as an important negative regulator of the STAT3 signaling pathway.20 As shown in Figures 4C and 5A, knockdown of RNF6 inhibited the phosphorylation of STAT3, but upregulated the expression levels of SHP-1. In addition, the mRNA levels of SHP-1 were not affected by knockdown of RNF6 (Figure 5B). To further confirm this result, NCI-N87 cells were transfected with increased concentrations of Myc-RNF6 plasmids, and we found that the protein level of SHP-1 was significantly downregulated in a dose-dependent manner (Figure 5C), but the mRNA level was not affected (Figure 5D). Furthermore, RNF6, the E3 ligase, induced the polyubiquitination of SHP-1 protein (Figure 5E). These results suggested that RNF6 regulated the stability of SHP-1 protein by affecting its polyubiquitination in gastric cancer cells.
Discussion
As shown by the successful application of bortezomib in the treatment of hematological malignancies, the ubiquitin–proteasome system (UPS) has been a useful target for antitumor drug discovery.21–23 But many patients developed resistance to bortezomib, and one potential strategy to combat bortezomib resistance is to target upstream components of the UPS, such as E3 ligase.24,25 Therefore, investigating the function of E3 ligases in tumorigenesis is significant for tumor therapy in clinical practice.
Recently, the function of E3 ligase RNF6 in tumorigenesis and tumor development has generated much attention. However, the function of RNF6 in gastric cancer is still unknown. In this study, RNF6 was demonstrated to be an oncogene in gastric cancer. RNF6 was highly expressed in gastric cancer tissues and cell lines (Figure 1), and knockdown of RNF6 induced gastric cancer cell growth inhibition and apoptosis (Figures 2 and 3). Interestingly, a recent study reported that saponins, the amphipathic glycosides found in traditional Chinese medicines, displayed potent antileukemia activity by suppressing RNF6 expression along with inactivated AKT/mTOR signaling.26 In contrast, RNF6 was also believed to be a tumor suppressor in esophageal squamous cell carcinomas because of its chromosomal location and somatic mutations.5 Together with these reports, we believed that RNF6 was a double-faceted gene in different cell types and could be as a target for specific tumor therapy.
STAT3 is an oncogenic transcription factor, and constitutive STAT3 activation induces tumorigenesis and commonly suggests poor prognosis in many tumors, including gastric cancer.27,28 STAT3 has been considered as an ideal drug target for various cancer treatments.29,30 In this study, we identified that RNF6 regulated STAT3 activation by affecting the polyubiquitination of SHP-1, which was a negative regulator of STAT3 signaling, and RNF6-SHP-1-STAT3 pathway could be a promising target for gastric cancer treatment (Figures 4 and 5).
DOX is a common chemotherapeutic drug in the treatment of gastric cancer, but patients frequently develop resistance to it partly because of overactivated STAT3.31,32 As described earlier, RNF6 enhanced the activation of STAT3, therefore we wanted to assess whether suppressing RNF6 could enhance the efficacy of DOX in gastric cancer cells. As shown in Figure 3C and D, knockdown of RNF6 enhanced the cytotoxic activity of DOX against gastric cancer cells, which showed potential clinical relevance and was meaningful for drug resistance therapy. Therefore, targeting RNF6 could be an effective strategy for gastric cancer treatment in the future.
Conclusion
This study demonstrated that RNF6 triggered STAT3 signaling, thus promoting the growth of gastric cancer cells. This study suggests that targeting RNF6-SHP-1-STAT3 pathway can be a promising approach for gastric cancer therapy in the future.
Ethics approval and consent to participate
This study was approved by the review board and ethical committee of The Affiliated Huaian No 1 People’s Hospital of Nanjing Medical University, and each patient provided written informed consent to donate tissues for this study after clinical procedures.
Acknowledgments
This study was supported by the General Project of Science and Technology Development Fund of Nanjing Medical University (grant no 2013NJMU225). The authors thank Dr Jerry Xu for his discussion and design of this project.
Disclosure
The authors report no conflicts of interest in this work.
References
Li S, Zhuang Z, Wu T, et al. Nicotinamide nucleotide transhydrogenase-mediated redox homeostasis promotes tumor growth and metastasis in gastric cancer. Redox Biol. 2018;18:246–255. | ||
Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Chang P, Wang F, Li Y. Hsa_circ_0000673 is down-regulated in gastric cancer and inhibits the proliferation and invasion of tumor cells by targeting miR-532-5p. Biosci Rep. 2018:BSR20180538. | ||
Tursun B, Schlüter A, Peters MA, et al. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 2005;19(19):2307–2319. | ||
Lo HS, Hu N, Gere S, et al. Identification of somatic mutations of the RNF6 gene in human esophageal squamous cell carcinoma. Cancer Res. 2002;62(15):4191–4193. | ||
Liu L, Zhang Y, Wong CC, et al. RNF6 Promotes Colorectal Cancer by Activating the Wnt/β-Catenin Pathway via Ubiquitination of TLE3. Cancer Res. 2018;78(8):1958–1971. | ||
Liang Q, Ma D, Zhu X, et al. RING-Finger Protein 6 Amplification Activates JAK/STAT3 Pathway by Modifying SHP-1 Ubiquitylation and Associates with Poor Outcome in Colorectal Cancer. Clin Cancer Res. 2018;24(6):1473–1485. | ||
Xu X, Han K, Tang X, et al. The Ring Finger Protein RNF6 Induces Leukemia Cell Proliferation as a Direct Target of Pre-B-cell Leukemia Homeobox 1. J Biol Chem. 2016;291(18):9617–9628. | ||
Zeng Y, Xu X, Wang S, et al. Ring finger protein 6 promotes breast cancer cell proliferation by stabilizing estrogen receptor alpha. Oncotarget. 2017;8(12):20103–20112. | ||
Xu K, Shimelis H, Linn DE, et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell. 2009;15(4):270–282. | ||
Xu X, Wang J, Han K, et al. Antimalarial drug mefloquine inhibits nuclear factor kappa B signaling and induces apoptosis in colorectal cancer cells. Cancer Sci. 2018;109(4):1220–1229. | ||
Xu X, Zhang J, Han K, et al. Natural pesticide dihydrorotenone arrests human plasma cancer cells at the G0/G1 phase of the cell cycle. J Biochem Mol Toxicol. 2014;28(5):232–238. | ||
Han K, Xu X, Chen G, et al. Identification of a promising PI3K inhibitor for the treatment of multiple myeloma through the structural optimization. J Hematol Oncol. 2014;7:9. | ||
Chen G, Xu X, Tong J, et al. Ubiquitination of the transcription factor c-MAF is mediated by multiple lysine residues. Int J Biochem Cell Biol. 2014;57:157–166. | ||
Han K, Xu X, Xu Z, et al. SC06, a novel small molecule compound, displays preclinical activity against multiple myeloma by disrupting the mTOR signaling pathway. Sci Rep. 2015;5:12809. | ||
Xu X, Han K, Zhu J, et al. An inhibitor of cholesterol absorption displays anti-myeloma activity by targeting the JAK2-STAT3 signaling pathway. Oncotarget. 2016;7(46):75539–75550. | ||
Ji ZP, Qiang L, Zhang JL. Transcription activated p73-modulated cyclin D1 expression leads to doxorubicin resistance in gastric cancer. Exp Ther Med. 2018;15(2):1831–1838. | ||
Kang X, Li M, Zhu H, et al. DUSP4 promotes doxorubicin resistance in gastric cancer through epithelial-mesenchymal transition. Oncotarget. 2017;8(55):94028–94039. | ||
Xu J, Liu D, Niu H, et al. Resveratrol reverses Doxorubicin resistance by inhibiting epithelial-mesenchymal transition (EMT) through modulating PTEN/Akt signaling pathway in gastric cancer. J Exp Clin Cancer Res. 2017;36(1):19. | ||
Han Y, Amin HM, Franko B, et al. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood. 2006;108(8):2796–2803. | ||
Morrow JK, Lin HK, Sun SC, Zhang S. Targeting ubiquitination for cancer therapies. Future Med Chem. 2015;7(17):2333–2350. | ||
Huang Z, Wu Y, Zhou X, et al. Efficacy of therapy with bortezomib in solid tumors: a review based on 32 clinical trials. Future Oncol. 2014;10(10):1795–1807. | ||
Wallington-Beddoe CT, Sobieraj-Teague M, Kuss BJ, Pitson SM. Resistance to proteasome inhibitors and other targeted therapies in myeloma. Br J Haematol. 2018;182(1):11–28. | ||
Farshi P, Deshmukh RR, Nwankwo JO, et al. Deubiquitinases (DUBs) and DUB inhibitors: a patent review. Expert Opin Ther Pat. 2015;25(10):1191–1208. | ||
Malek E, Abdel-Malek MA, Jagannathan S, et al. Pharmacogenomics and chemical library screens reveal a novel SCFSKP2 inhibitor that overcomes Bortezomib resistance in multiple myeloma. Leukemia. 2017;31(3):645–653. | ||
Lu Q, He Y, Wang Y, et al. Saponins From Paris forrestii (Takht.) H Li Display Potent Activity Against Acute Myeloid Leukemia by Suppressing the RNF6/AKT/mTOR Signaling Pathway. Front Pharmacol. 2018;9:673. | ||
Cafferkey C, Chau I. Novel STAT 3 inhibitors for treating gastric cancer. Expert Opin Investig Drugs. 2016;25(9):1023–1031. | ||
Tong M, Wang J, Jiang N, Pan H, Li D. Correlation between p-STAT3 overexpression and prognosis in lung cancer: A systematic review and meta-analysis. PLoS One. 2017;12(8):e0182282. | ||
Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. | ||
Kitamura H, Ohno Y, Toyoshima Y, et al. Interleukin-6/STAT3 signaling as a promising target to improve the efficacy of cancer immunotherapy. Cancer Sci. 2017;108(10):1947–1952. | ||
Vanklompenberg MK, Leyden E, Arnason AH, et al. APC loss in breast cancer leads to doxorubicin resistance via STAT3 activation. Oncotarget. 2017;8(61):102868–102879. | ||
Zhang ZL, Jiang QC, Wang SR. Schisandrin A reverses doxorubicin-resistant human breast cancer cell line by the inhibition of P65 and Stat3 phosphorylation. Breast Cancer. 2018;25(2):233–242. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.