Back to Journals » Cancer Management and Research » Volume 12
Knockdown of PVT1 Suppresses Colorectal Cancer Progression by Regulating MiR-106b-5p/FJX1 Axis
Authors Liu F, Wu R, Guan L, Tang X
Received 29 April 2020
Accepted for publication 7 August 2020
Published 22 September 2020 Volume 2020:12 Pages 8773—8785
DOI https://doi.org/10.2147/CMAR.S260537
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Chien-Feng Li
Fang Liu,1 Rong Wu,2 Lina Guan,3 Xuegui Tang1
1Anorectal Department of Integrated Traditional Chinese and Western Medicine, Affiliated Hospital of North Sichuan Medical College, Nanchong, Sichuan, People’s Republic of China; 2Department of Clinical Medicine of Combination of Chinese and Western Medicine, North Sichuan Medical College, Nanchong, Sichuan, People’s Republic of China; 3Institute of Anorectal Diseases, North Sichuan Medical College, Nanchong, Sichuan, People’s Republic of China
Correspondence: Xuegui Tang Anorectal Department of Integrated Traditional Chinese and Western Medicine
Affiliated Hospital of North Sichuan Medical College, No. 1, Maoyuan South Road, Nanchong 637000, Sichuan, People’s Republic of China
Tel +86-0817-2262120
Email [email protected]
Purpose: Long non-coding RNA plasmacytoma variant translocation 1 (PVT1) has been revealed to involve in the progression of CRC. However, the precise mechanisms of PVT1 in action remain unclear.
Methods: The expression of PVT1, microRNA-106b-5p (miR-106b-5p) and four jointed box 1 (FJX1) was measured using quantitative real-time polymerase chain reaction (qRT-PCR) or Western blot, respectively. Cell proliferation was investigated by 3-(4,5)-dimethylthiahiazo (−z-y1)-3,5-di-phenytetrazoliumromide assay. Transwell assay was used to determine cell migration and invasion. The correlation between miR-106b-5p and PVT1 or FJX1 was confirmed using luciferase reporter assay. The effects of PVT1 in vivo were assessed using mice xenograft model.
Results: PVT1 was up-regulated in CRC tissues and cell lines, especially in CRC tissues with high-grade, and highly expressed PVT1 predicted worse prognosis. Functional experiments demonstrated that PVT1 deletion inhibited CRC cell proliferation, migration and invasion in vitro and suppressed tumor growth in vivo. MiR-106b-5p was confirmed to be a target of PVT1, and inhibition of miR-106b-5p reversed the inhibitory effects of PVT1 knockdown on CRC cell malignant phenotypes. In addition, we found miR-106b-5p directly targeted FJX1, and miR-106b-5p-mediated inhibition on CRC cell proliferation, migration and invasion was attenuated by FJX1 up-regulation. Importantly, it was also proved that PVT1 could indirectly regulate FJX1 expression via targeting miR-106b-5p.
Conclusion: Knockdown of PVT1 impaired cell proliferation, migration and invasion in CRCs via regulating miR-106b-5p/FJX1 axis, which provided a novel insight into the development of therapeutic strategies for CRC patients.
Keywords: colorectal cancer, PVT1, miR-106b-5p, FJX1, metastasis, apoptosis
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-related mortality worldwide and has grown rapidly in China in recent years.1 Despite the development in systemic therapy and liver-directed treatments, about 86% of patients with advanced disease still die within 5 years after diagnosed.2 Therefore, a better understanding on molecular mechanisms and genetic alterations underlying CRC to find out the novel therapeutic strategies in treating CRC is of great importance.
Long non-coding RNAs (LncRNAs) are a class of non-coding ribonucleic acids (RNAs), which are longer than 200 nts in size. LncRNAs are recognized as vital participants in epigenetic regulation, gene transcription regulation, and other essential biological processes, which are involved in tumorigenesis.3,4 LncRNA plasmacytoma variant translocation 1 (PVT1), encoded by the human PVT1 gene, is located on chromosome 8q24.21 proximity to c-Myc, which implicates in the susceptibility and progression of cancer.5 PVT1 gene displayed frequent amplification at genome level and concordant transcriptional up-regulation in different malignancies.6,7 Increasing evidence has revealed the carcinogenic effects of PVT1 on various tumors, such as non-small-cell lung cancer, hepatocellular cancer, leukemia, ovarian and breast cancer.8–11 In CRC, several studies suggested that PVT1 functioned as an oncogene to promote CRC tumorigenesis through regulating cell biological behavior.12,13 However, the precise mechanisms of PVT1 in action remain unclear.
The lncRNA-miRNA-mRNA network has been revealed to be a vital step toward understanding where lncRNAs fit into the biological processes behind tumor development.11 Up to now, various microRNAs (miRNAs) were found to directly involve in PVT1-mediated tumorigenesis. For example, the amplification of PVT1 promoted ovarian cancer cell proliferation by binding to miR-140;14 PVT1 promoted cell proliferation, invasion and epithelial–mesenchymal transition (EMT) in renal cell carcinoma via mediating the down-regulation of miR-16-5p.15 MiR-106b-5p, a member of miR-106b ~ 25 clusters, has also been identified to be up-regulated in CRC tissues and implicate in the invasion and metastasis of CRC cells.16,17 Four jointed box 1 (FJX1) is a notch-inducible secreted ligand, which is homologous to the Drosophila four-jointed (fj) gene.18 Although the exact biological function of FJX1 in human cancers remains unclear, studies of different species indicate that FJX1 participates in carcinogenesis and FJX1 gene amplification as well as subsequent mRNA expression have been found, besides, FJX1 has also been considered as a candidate for regulation of angiogenesis in several human cancers, including CRC.19–21 However, whether specific crosstalk existed between PVT1 and FJX1 through competition for miR-106b-5p binding in CRC is unclear.
In the present study, we focused on investigating the role and molecular mechanism of PVT1 in CRC cell proliferation and metastasis, and further explored the potential regulatory relationship among PVT1, FJX1 and miR-106b-5p in the progression of CRC in vivo and vitro, which might provide a potential therapeutic target for CRC treatment.
Methods
Patients and Specimens
A total of 62 primary CRC tissues (31 cases of grade I–II, 31 cases of grade III–IV according to WHO grading system) and corresponding non-tumor tissues were obtained from CRC patients who underwent surgery at Affiliated Hospital of North Sichuan Medical College. All CRC specimens were diagnosed by two independent pathologists and did not receive any preoperative treatment. Specimens were immediately stored at −80°C until further study.
The clinical features, including age, gender, tumor size, TNM stages, lymphatic metastasis, vascular invasion, and distant metastasis were collected from the recruited cases. Follow-up was conducted regularly every 3 months in the first 2 years after surgery, and reduced to once every 6 months from the third year. The last follow-up was carried out in July 2017. The study was approved by the Ethics Committee of Affiliated Hospital of North Sichuan Medical College and written informed consent has been obtained from all subjects.
Cell Culture and Transfection
Colorectal carcinoma cell lines HCT116 and HT29 were purchased from the American Type Culture Collection (ATCC; Rockville, MD, USA). Human NCM460 colonocytes were obtained from Shanghai Cell Bank of the Chinese Academy of Science (Shanghai, China). HT-29 and HCT-116 cells were maintained in McCoy’s 5A medium (HyClone, Logan, UT). NCM460 were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Carlsbad, CA, USA). All media were supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Gibco). All cells were incubated at 37°C with 5% CO2.
The short hairpin RNA (shRNA) targeting PVT1 (sh-PVT1) and shRNA scramble control (sh-NC), pcDNA3.1 empty vector (vector), pcDNA3.1-PVT1 overexpression vector (PVT1), pcDNA3.1-FJX1 overexpression vector (FJX1) were synthesized by Genepharma (Shanghai, China). The miR-106b-5p mimic (miR-106b-5p), mimic negative control (miR-NC), miR-106b-5p inhibitor (anti-miR-106b-5p) and inhibitor negative control (anti-NC) were purchased from RIBOBIO (Guangzhou, China). All oligonucleotides or vectors were transfected into HCT116 and HT29 cells by using LipofectamineTM 2000 reagent (Invitrogen, Carlsbad, CA, USA), respectively. Then, cells were harvested for 48 h for the subsequent study.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from CRC tissues and the treated HCT116 and HT29 cells using TRIzol reagent (Invitrogen) according to the manufacturer’s introductions. The cDNA was synthesized from extracted RNA by using the PrimeScript Reverse Transcription Kit (Invitrogen). Then, qRT-PCR was performed using the SYBR Green PCR Master Mix kit (Takara, Dalian, China) on a PRISM® 7300 system (Applied Biosystems, Foster City, CA, USA). U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as endogenous control and the 2−ΔΔCt method was applied to calculate the relative expression levels. The special primers for miR-106b-5p or U6 were purchased from Qiagen (Hilden, Germany) and the following primers were listed: PVT1 forward 5ʹ-TGAGAACTGTCCTTACGTGACC-3ʹ and reverse 5ʹ-AGAGCACCA AGACTG GCTCT-3ʹ; GAPDH forward 5ʹ-CCGGGAAACTGTGGCGTGATGG-3ʹ and reverse 5ʹ-AGGTGGAGGAGTGGGTGTCGCTGTT-3ʹ; FJX1 forward 5ʹ-CGTGC TGGCACAGTAAAGAA-3ʹ and reverse 5ʹ-CAAAGTTCTGGGAGGACG-3ʹ.
Cell Proliferation Ability
Cell proliferation ability was determined using the 3-(4,5)-dimethylthiahiazo (−z-y1)-3,5-di-phenytetrazoliumromide (MTT; Beyotime, Shanghai, China) assay. In brief, transfected cells were seeded into 96-well plate with a density of 5 × 103 cells/well and incubated for 0 h, 24 h, 48 h or 72 h. At different time points, each well was added with 20 μL of MTT solution for 4 h, followed by the addition of DMSO to resolve the generated formazan. Finally, the absorbance was measured at 490 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
Cell Migration and Invasion Assay
Cell migration and invasion abilities were measured using transwell assay. For cell migration assay, transfected HCT116 and HT29 cells were resuspended and adjusted to the cell concentration to 1×105/mL. Then, cells were seeded on the upper transwell chamber with a noncoated membrane in McCoy’s 5A medium without serum, and complete medium supplemented with 10% FBS was added to the lower chamber as a chemoattractant. After incubation for 24 h, the cells in the upper membrane were removed with a dry cotton swab, and cells located on the lower surface of the membrane fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The cells number of five randomly selected fields were counted under a microscope (Olympus, Tokyo, Japan). For the invasion assay, the method of measurement was similar to the process of cell migration, except that upper transwell chambers were pre-coated with Matrigel (BD, San Jose, CA, USA) for 1 h at 37°C.
Luciferase Reporter Assay
The PVT1 mRNA and FJX1 3ʹ-UTR containing wild-type (WT) or mutant (MUT) binding sequence of miR-106b-5p were cloned into the psiCHECKTM-2 luciferase plasmid (Promega, Shanghai, China), respectively. Then, HCT116 and HT29 cells were cultured in 96-well plates and co-transfected with 20 pmol of the miR-106b-5p mimic or miR-NC mimic, and 50 ng of luciferase reporters or 50 ng of a control luciferase plasmid using LipofectamineTM 2000. After transfection for 48 h, a dual-luciferase assay kit (Promega) was used to investigate the luciferase activity according to the manufacturer’s instructions.
Western Blot Assay
Western blot assays were carried out as previously described.22 Cells were lysed with RIPA lysis buffer (Beyotime, Shanghai, China) and quantified by using a bicinchoninic acid (BCA) protein assay kit (Beyotime) according to the manufacturer’s instructions. Approximately 25 μg of extracted proteins were subjected to 10% SDS-polyacrylamide gels electrophoresis, transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA). Interaction with primary antibodies FJX1 and GAPDH, and followed by incubation with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody. Immunoreactive signals were visualized using Enhanced Chemiluminescence (ECL).
Murine Xenograft Assay
BALB/c-nu/nu mice (5-week-old) were bought from Vital River Laboratory Animal Technology (Beijing, China). All research involving animals complied with protocols was approved by the Animal Research Committee of Affiliated Hospital of North Sichuan Medical College and was carried out according to the guidelines of the National Animal Care and Ethics Institution. HCT116 cells (1 × 106) transfected with the lentivirus containing short hairpin RNA targeting PVT1 (sh-PVT1) or negative control (sh-NC) were subcutaneously transplanted into the mice and the tumor volumes were detected and calculated 0 d, 7 d, 14 d, 21 d and 28 d after injection. After 30 d, all mice were sacrificed and tumor samples were weighted and used for further molecular analysis.
Statistical Analysis
All data were expressed as the mean ± standard deviation (SD). The differences between groups were analyzed with Student’s t-test using the GraphPad Prism 7 (GraphPad Inc., San Diego, CA, USA). The correlation between PVT1 and clinicopathological features of the CRC patients was analyzed with the chi-square test. Kaplan–Meier survival curves were plotted and the difference in survival between two groups was analyzed by the Log rank test. The linear relationship between miR-106b-5p and PVT1 or FJX1 mRNA was determined via Pearson’s correlation analysis. P<0.05 indicated statistically significant.
Results
PVT1 is Up-Regulated in CRC Tissues and Cell Lines and Highly Expressed PVT1 Predicts Poor Prognosis
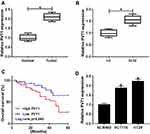
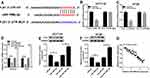
The mRNA expression of PVT1 was detected in CRC tissues and adjacent noncancerous tissues using qRT-PCR, results showed PVT1 was greatly elevated in CRC tissues compared with normal tissues (Figure 1A), and the expression of PVT1 in the high-grade CRC tissues (grade III–IV, n=31) was significantly higher than that in the low-grade CRC tissues (grade I–II, n=31) (Figure 1B). Additionally, the correlation between the expression of PVT1 and clinicopathological features in 62 CRC patients was analyzed (Table 1). The results showed that PVT1 expression was closely correlated to CRC tumor size, TNM stages, lymphatic metastasis, and distant metastasis while not associated with age, gender and vascular invasion. Moreover, the correlation between the PVT1 expression and overall survival (OS) was analyzed using Kaplan–Meier analysis and Log rank test in 62 CRC patients and the data suggested that highly expressed PVT1 predicted worse prognosis (Figure 1C). Subsequently, PVT1 mRNA expression in CRC cells was measured, qRT-PCR analysis showed compared with the normal human NCM460 colonocytes, PVT1 expression was also up-regulated in CRC cell lines including HCT116 and HT29 (Figure 1D). Thus, all the results suggested PVT1 was increased in CRC and might be associated with the progression of CRC.
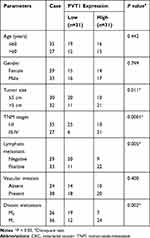 |
Table 1 Correlation Between PVT1 Expression and Clinical Clinicopathological Parameters of CRC Patients (n=62) |
PVT1 Deletion Inhibits Cell Proliferation, Migration and Invasion Abilities in CRC
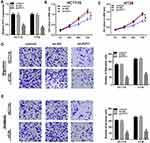
To explore the biological function of PVT1 in CRC, CRC cells were transfected with sh-NC or sh-PVT1 and then the transfection efficiency was measured by qRT-PCR with the results of decreased PVT1 mRNA expression in HCT116 and HT29 cells transfected with sh-PVT1 (Figure 2A). Then, cell proliferation activity was measured using MTT assay and results showed that knockdown of PVT1 suppressed the proliferation of HCT116 and HT29 cells (Figure 2B and C). Subsequently, transwell assay exhibited that transfection with PVT1 shRNA markedly repressed the migration and invasion abilities of HCT116 and HT29 cells (Figure 2D and E). Besides that, HCT116 and HT29 cells also were transfected with PVT1 or Vector to conduct gain-of-function research. As expected, PVT1 transfection significantly increased the level of PVT1 in HCT116 and HT29 cells (Fig. S1 A), then MTT assay showed PVT1 overexpression remarkably promoted the proliferation of HCT116 and HT29 cells (Fig. S1 B, C). These results indicated that PVT1 silencing inhibited cell proliferation, migration and invasion in CRC.
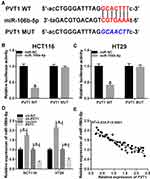
PVT1 Directly Targets MiR-106b-5p and Negatively Regulates Its Expression in CRC Cells
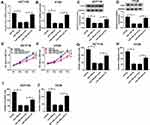
Through the prediction of Starbase program, miR-106b-5p was found that might be a potential target of PVT1 with a putative binding site (Figure 3A). To verify this prediction, luciferase reporter assay was performed and we found miR-106b-5p mimic significantly suppressed the luciferase activity of PVT1-WT, while there was no apparent change in PVT1-MUT reporter after overexpression miR-106b-5p in HCT116 and HT29 cells (Figure 3B and C). Then, qRT-PCR was used to examine the effects of PVT1 on miR-106b-5p mRNA expression in HCT116 and HT29 cells, the results suggested that PVT1 silencing promoted miR-106b-5p expression, while PVT1 overexpression inhibited miR-106b-5p expression in HCT116 and HT29 cells (Figure 3D). Furthermore, a negative correlation between miR-106b-5p and PVT1 mRNA expression was investigated in CRC (Figure 3E). Taken together, we confirmed that PVT1 directly bound to miR-106b-5p and negatively regulated its expression in CRC cells.
PVT1 Silencing Inhibits the Proliferation, Migration and Invasion Abilities of CRC Cells Through Regulating MiR-106b-5p Expression
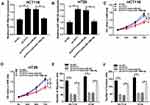
To investigate whether the effect of PVT1 on CRC cell biological behaviors was mediated by miR-106b-5p, HCT116 and HT29 cells were transfected with sh-NC, sh-PVT1, sh-PVT1 + anti-miR-NC or sh-PVT1 + anti-miR-106b-5p to conduct rescue assay. After transfection, we found the introduction of miR-106b-5p inhibitor reduced the expression of miR-106b-5p in sh-PVT1-transfected HCT116 and HT29 cells (Figure 4A and B). After that, results from MTT and transwell assays indicated that miR-106b-5p inhibitor could reverse PVT1 deletion-induced inhibitory effects on the proliferation, migration and invasion of HCT116 and HT29 cells (Figure 4CF) (images for cell migration and invasion, Additional file 1: Fig. S2). Thus, we verified that knockdown of PVT1 suppressed CRC progression via regulating miR-106b-5p expression.
MiR-106b-5p Directly Targets FJX1 and Suppresses FJX1 Expression
According to the prediction of Targetscan program, we found that the 3ʹUTR of FJX1 contained the potential binding sites for miR-106b-5p (Figure 5A). FJX1 might be a target of miR-106b-5p. Subsequently, to validate the prediction, we conducted luciferase reporter assay and results showed miR-106b-5p overexpression reduced the luciferase activities of the FJX1-WT reporter vector but not FJX1-MUT reporter vector in HCT116 and HT29 cells (Figure 5B and C). In the meanwhile, the expression of FJX1 was measured using qRT-PCR and Western blot, and we demonstrated that miR-106b-5p up-regulation inhibited FJX1 expression while miR-106b-5p down-regulation promoted FJX1 expression, whether the mRNA or protein expression, in HCT116 and HT29 cells (Figure 5DF). Besides that co-expression analysis exhibited a perfect negative correlation between FJX1 and miR-106b-5p mRNA expression in CRC (Figure 5G). Therefore, all the data suggested that FJX1 was a target of miR-106b-5p and was negatively regulated by miR-106b-5p in CRC cells.
Overexpressed MiR-106b-5p Suppresses the Proliferation, Migration and Invasion Abilities of CRC Cells via Regulating FJX1 Expression
We then determined whether miR-106b-5p regulated cell malignant phenotypes via the FJX1. HCT116 and HT29 cells were transfected with miR-NC, miR-106b-5p, miR-106b-5p + FJX1, or miR-106b-5p + control vector, and then qRT-PCR and Western blot analysis revealed the expression of FJX1 at mRNA and protein levels were reduced by miR-106b-5p re-expression, while this reduction was rescued by FJX1 overexpression (Figure 6AD). Thereafter, the proliferation, migration and invasion abilities of CRC cells were detected. Results of MTT assay showed that miR-106b-5p mimic inhibited the proliferation activity, while FJX1 up-regulation could restore this effect in HCT116 and HT29 cells (Figure 6E and F). Then, transwell assay showed that FJX1 transfection attenuated miR-106b-5p re-expression-induced suppression of the migration and invasion of HCT116 and HT29 cells (Figure 6GJ) (images for cell migration and invasion, Additional file 1: Fig. S3). Altogether, these results suggested that overexpressed miR-106b-5p could suppress the proliferation, migration and invasion abilities of CRC cells via regulating FJX1 inhibition.
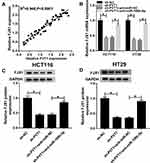
PVT1 Regulates FJX1 Expression via MiR-106b-5p
A perfect positive correlation between FJX1 and PVT1 mRNA expression was determined in CRC tissues (Figure 7A), indicating the potential regulatory relationship among FJX1, miR-106b-5p and PVT1. Afterward, co-expression analysis using HCT116 and HT29 cell showed that the expression of FJX1, whether mRNA or protein, was inhibited by PVT1 deletion, while this condition was reversed by miR-106-5p inhibitor (Figure 7BD), suggesting a feedback loop and a complex regulatory network among PVT1/miR-106b-5p/FJX1 axis.
Knockdown of PVT1 Suppresses CRC Tumor Growth in vivo
To identify the effect of PVT1 on CRC oncogenesis in vivo, HCT116 cells, stably transfected with sh-PVT1 and sh-NC, were subcutaneously injected into nude mice to establish mice xenograft model. As exhibited in Figure 8A, B, PVT1 silencing significantly suppressed the growth of CRC tumors, reflected by the reduction of tumor volume and weight. Furthermore, molecular analyses were conducted in harvested tumor tissues, qRT-PCR analysis revealed that sh-PVT1 transfection decreased the mRNA expression of PVT1 and FJX1 in tumors, while the mRNA expression of miR-106b-5p was up-regulated; besides that, Western blot analysis exhibited the protein level of also was down-regulated in tumor tissues form sh-PVT1-transfected mice (Figure 8CF), indicating PVT1 might regulate FJX1 expression by miR-106b-5p in vivo, which was consistent with the results in vitro. Thus, we demonstrated that knockdown of PVT1 might suppress CRC tumor growth in vivo via regulating miR-106b-5p/FJX1 axis.
Discussion
Numerous studies have reported that lncRNAs are involved in various aspects of cell biology and potentially contribute to cancer development. The roles of lncRNAs as drivers of oncogenic or anticancer function have been demonstrated in diverse tumors.23 In CRC, many lncRNAs have also been identified to be closely associated with CRC progression by regulating cell biological processes.24 For example, lncRNA CPS1-IT1 suppressed cell proliferation, invasion and metastasis in CRC;25 LncRNA-UCA1 enhanced cell proliferation and 5-fluorouracil resistance in CRC by inhibiting miR-204-5p;26 LncRNA CRNDE promoted CRC cell proliferation and chemoresistance via regulating miR-181a-5p/Wnt/β-catenin signaling.27 All the studies revealed that lncRNAs play important roles in CRC development.
PVT1 has been identified to be elevated in CRC tissues and correlated with poor prognoses in patients suffering from CRC.28,29 In the current study, we discovered that PVT1 was up-regulated in CRC tissues, especially in the high-grade CRC tissues, and its high expression predicted worse prognosis which was consistent with previous studies. Additionally, PVT1 expression was closely correlated to CRC tumor size, TNM stages, lymphatic metastasis, and distant metastasis, while not associated with age, gender and vascular invasion. Besides, PVT1 expression was also increased in CRC cell lines, functional experiments showed PVT1 deletion inhibited the proliferation, migration and invasion abilities of CRC cells in vitro. All the findings indicated the potential regulatory role of PVT1 in the development of CRC.
LncRNAs can act as “miRNA sponges” and sequester miRNAs to inactivate these small regulatory RNAs and then to reduce the expression of miRNA target genes.24 In the prior studies, Shang et al found PVT1 acted as a competing endogenous RNA (ceRNA) of miR-214-3p to promote cell proliferation and invasion in CRC;22 Zhang et al demonstrated that PVT1 promoted colon cancer progression via endogenous sponging miR-26b.30 In addition, PVT1 also was revealed to regulate CRC tumor development by sponging miR-455.13 In the meanwhile, Zhuang et al indicated MALAT1 sponged miR-106b-5p to promote the invasion and metastasis of CRC via SLAIN2 enhanced microtubules mobility.17 In this study, we confirmed that PVT1 directly interacted with miR-106b-5p and negatively regulated miR-106b-5p expression in CRC cells. Functional assay showed overexpressed miR-106b-5p functioned as a tumor suppressor in CRC cell progression by inhibiting cell proliferation and metastasis. Subsequently, rescue assay showed that down-regulated miR-106b-5p could reverse PVT1 silencing-mediated inhibition on CRC tumorigenesis.
MiRNAs regulate post-transcriptional expression of genes through binding to the 3ʹ-UTR of their target mRNAs.31 In the present study, using bioinformatics tools, FJX1 was confirmed to be a target of miR-106b-5p and was negatively regulated by miR-106b-5p. Then, rescue assay showed that FJX1 vector transfection attenuated miR-106b-5p overexpression-induced suppression of cell proliferation, migration and invasion abilities in CRC. Moreover, a positive correlation between PVT1 and FJX1 was illustrated and co-expression analysis suggested PVT1 could regulate FJX1 expression via targeting miR-106b-5p expression in vitro. Thus, a feedback loop consisting of PVT1/miR-106b-5p/FJX1 in CRC progression in vitro was identified. Afterward, in order to consolidate our findings, the effects of PVT1 in vivo were assessed using mice xenograft model, the results showed that knockdown of PVT1 expression suppressed the growth of CRC tumor in vivo. Furthermore, it was also observed that PVT1 could also regulate FJX1 expression by competitive binding to miR-106b-5p in vivo, indicating the regulatory effects of PVT1/miR-106b-5p/FJX1 axis in CRC development.
Conclusion
In conclusion, PVT1 was found to promote cell proliferation, migration and invasion abilities in CRC via regulating miR-106b-5p/FJX1 axis. We firstly identified the regulatory network of PVT1/miR-106b-5p/FJX1 axis in CRC progression, which provided a potential therapeutic target for CRC treatment.
Acknowledgments
The authors would like to thank the participants in this study.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi:10.3322/caac.21395
2. Gu MJ, Huang QC, Bao CZ, et al. Attributable causes of colorectal cancer in China. BMC Cancer. 2018;18:38. doi:10.1186/s12885-017-3968-z
3. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi:10.1534/genetics.112.146704
4. Gudenas BL, Wang J, Kuang SZ, et al. Genomic data mining for functional annotation of human long noncoding RNAs. J Zhejiang Univ Sci B. 2019;20:476–487. doi:10.1631/jzus.B1900162
5. Tseng YY, Moriarity BS, Gong W, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi:10.1038/nature13311
6. Adhikary J, Chakraborty S, Dalal S, et al. Circular PVT1: an oncogenic non-coding RNA with emerging clinical importance. J Clin Pathol. 2019;72:513–519. doi:10.1136/jclinpath-2019-205891
7. Wang F, Yuan JH, Wang SB, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi:10.1002/hep.27239
8. Wang D, Hu Y. Long non-coding RNA PVT1 competitively binds MicroRNA-424-5p to regulate CARM1 in radiosensitivity of non-small-cell lung cancer. Mol Ther Nucleic Acids. 2018;16:130–140. doi:10.1016/j.omtn.2018.12.006
9. Guo J, Hao C, Wang C, et al. Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int. 2018;18:98. doi:10.1186/s12935-018-0582-3
10. El-Khazragy N, Elayat W, Matbouly S, et al. The prognostic significance of the long non-coding RNAs CCAT1, PVT1 in t (8;21) associated acute myeloid leukemia. Gene. 2019;707:172–177. doi:10.1016/j.gene.2019.03.055
11. Guan Y, Kuo WL, Stilwell JL, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi:10.1158/1078-0432.CCR-06-2882
12. Guo K, Yao J, Yu Q, et al. The expression pattern of long non-coding RNA PVT1 in tumor tissues and in extracellular vesicles of colorectal cancer correlates with cancer progression. Tumor Biol. 2017;39:1010428317699122. doi:10.1177/1010428317699122
13. Chai J, Guo D, Ma W, et al. A feedback loop consisting of RUNX2/LncRNA-PVT1/miR-455 is involved in the progression of colorectal cancer. Am J Cancer Res. 2018;8:538–550.
14. Ding Y, Fang Q, Li Y, et al. Amplification of lncRNA PVT1 promotes ovarian cancer proliferation by binding to miR-140. Mamm Genome. 2019;30:217–225. doi:10.1007/s00335-019-09808-1
15. Ren Y, Huang W, Weng G, et al. LncRNA PVT1 promotes proliferation, invasion and epithelial–mesenchymal transition of renal cell carcinoma cells through downregulation of mir-16-5p. Onco Targets Ther. 2019;12:2563–2575. doi:10.2147/OTT.S190239
16. Ni S, Weng W, Xu M, et al. miR-106b-5p inhibits the invasion and metastasis of colorectal cancer by targeting CTSA. Onco Targets Ther. 2018;11:3835–3845. doi:10.2147/OTT.S172887
17. Zhuang M, Zhao S, Jiang Z, et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. E Bio Medicine. 2019;41:286–298. doi:10.1016/j.ebiom.2018.12.049
18. Afshar Y, Miele L, Fazleabas AT. Notch1 is regulated by chorionic gonadotropin and progesterone in endometrial stromal cells and modulates decidualization in primates. Endocrinology. 2012;153:2884–2896. doi:10.1210/en.2011-2122
19. Chai SJ, Ahmad Zabidi MM, Gan SP, et al. An Oncogenic role for four-jointed box 1 (FJX1) in nasopharyngeal carcinoma. Dis Markers. 2019;2019:3857853. doi:10.1155/2019/3857853
20. Snijders AM, Schmidt BL, Fridlyand J, et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi:10.1038/sj.onc.1208601
21. Al-Greene NT, Means AL, Lu P, et al. Four jointed box 1 promotes angiogenesis and is associated with poor patient survival in colorectal carcinoma. PLoS One. 2013;8:e69660. doi:10.1371/journal.pone.0069660
22. Shang AQ, Wang WW, Yang YB, et al. Knockdown of long non-coding RNA PVT1 suppresses cell proliferation and invasion of colorectal cancer via upregulation of microRNA-214-3p. Am J Physiol Gastrointest Liver Physiol. 2019;317:G222G232. doi:10.1152/ajpgi.00357.2018
23. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi:10.1158/2159-8290.cd-11-0209
24. Xie X, Tang B, Xiao YF, et al. Long non-coding RNAs in colorectal cancer. Oncotarget. 2016;7:5226–5239. doi:10.18632/oncotarget.6446
25. Zhang W, Yuan W, Song J, et al. LncRna CPS1-IT1 suppresses cell proliferation, invasion and metastasis in colorectal cancer. Cell Physiol Biochem. 2017;44:567–580. doi:10.1159/000485091
26. Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi:10.1038/srep23892
27. Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16:9. doi:10.1186/s12943-017-0583-1
28. Yu X, Zhao J, He Y. Long non-coding RNA PVT1 functions as an oncogene in human colon cancer through miR-30d-5p/RUNX2 axis. J BUON. 2018;23:48–54.
29. Fan H, Zhu JH, Yao XQ. Long non-coding RNA PVT1 as a novel potential biomarker for predicting the prognosis of colorectal cancer. Int J Biol Markers. 20181724600818777242. doi:10.1177/1724600818777242
30. Zhang R, Li J, Yan X, et al. Long noncoding RNA plasmacytoma variant translocation 1 (PVT1) promotes colon cancer progression via endogenous sponging miR-26b. Med Sci Monit. 2018;24:8685–8692. doi:10.12659/msm.910955
31. Zhao S, Han J, Zheng L, et al. MicroRNA-203 regulates growth and metastasis of breast cancer. Cell Physiol Biochem. 2015;37:35–42. doi:10.1159/000430331
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.