Back to Journals » Cancer Management and Research » Volume 12
Knockdown of MIR4435-2HG Suppresses the Proliferation, Migration and Invasion of Cervical Cancer Cells via Regulating the miR-128-3p/MSI2 Axis in vitro
Authors Wang R, Liu L, Jiao J, Gao D
Received 1 June 2020
Accepted for publication 2 August 2020
Published 22 September 2020 Volume 2020:12 Pages 8745—8756
DOI https://doi.org/10.2147/CMAR.S265545
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Ruijing Wang,1 Lun Liu,2 Jinwen Jiao,1 Dongmei Gao1
1Department of Gynecology, The Affiliated Hospital of Qingdao University, Qingdao City, Shandong Province 266555, People’s Republic of China; 2Department of Surgery, The Affiliated Hospital of Qingdao University, Qingdao City, Shandong Province 266555, People’s Republic of China
Correspondence: Dongmei Gao
Department of Gynecology, The Affiliated Hospital of Qingdao University, Qingdao City, Shandong Province 266555, People’s Republic of China
Tel +86- 0532- 82919505
Email [email protected]
Purpose: Long non-coding RNAs (lncRNAs) play major roles in the development of several cancers, including cervical cancer (CC). The purpose of the present study is to explore the regulatory mechanism of MIR4435-2HG on CC in vitro.
Patients and Methods: Fifty-nine pairs of CC tissues and adjacent normal tissues were collected from 59 patients by resection. The expression of lncRNA MIR4435-2HG, microRNA (miR)-128-3p and Musashi 2 (MSI2) in CC tissues and cells was detected by quantitative reverse-transcription PCR (qRT-PCR). The viability of CC cells was detected by 3-(4, 5-Dimethyl-2-Thiazolyl)-2, 5-Diphenyl-2-H-Tetrazolium Bromide (MTT) assay. The ability of migration and invasion in CC cells was measured by wound healing assay and transwell invasion assay, respectively. Starbase software and Targetscan software were utilized to predict the relationship between miR-128 and MIR4435-2HG/MSI2, respectively. The dual-luciferase reporter assay was used to confirm these interactions.
Results: LncRNA MIR4435-2HG expression was significantly up-regulated in CC tissues (P < 0.001) and cells (P < 0.01). Knockdown of MIR4435-2HG inhibited the proliferation, migration and invasion of CC cells (P < 0.01). MiR-128-3p was a target of MIR4435-2HG and was negatively modulated by MIR4435-2HG (P < 0.0001, r = − 0.6331). Up-regulation of miR-128-3p suppressed the proliferation, migration and invasion of CC cells (P < 0.01). In addition, MSI2 was the target gene of miR-128-3p and negatively regulated by miR-128-3p (P < 0.0001, r = − 0.4775). Both down-regulation of miR-128-3p and up-regulation of MSI2 reversed the inhibitory effects of MIR4435-2HG knockdown on the proliferation, migration and invasion of CC cells (P < 0.05).
Conclusion: MIR4435-2HG knockdown suppresses the proliferation, migration and invasion of CC cells through regulating the miR-128-3p/MSI2 axis, providing a possible therapeutic strategy for CC.
Keywords: cervical cancer, lncRNA MIR4435-2HG, miR-128-3p, MSI2
Introduction
Cervical cancer (CC), a type of malignant tumor that is common in females,1 accounts for a large proportion of tumor-resulted death globally.2,3 Currently, surgery, chemotherapy and radiotherapy are the primary therapeutic regimes for CC. Shockingly, only approximately 40% of CC patients can survive for more than 5 years.4,5 Therefore, Thus, understanding the mechanisms that underlie CC is critical for developing a new treatment strategy.
Long non-coding RNAs (lncRNAs) play major roles in tumorigenesis and progression of human cancers,6,7 including CC.8 LncRNA OIP5-AS1 has been revealed to promote the proliferation and inhibit cell apoptosis in CC.9 LncRNA PCAT6 has been reported to accelerate the progression of CC.10 LncRNA CAR10 expression is significantly up-regulated in CC tissues, whereas overexpression of CAR10 can promote the proliferation of CC cells.11 Additionally, the emerging evidences have displayed that lncRNA MIR4435-2HG is correlated with the occurrence and progression of many cancers.12–15 A previous study has showed that overexpression of lncRNA MIR4435-2HG increases the cell viability and migration in colon adenocarcinoma (COAD) cells, which eventually promotes the development of colorectal cancer (CRC) in mice.14 Another study has disclosed that MIR4435-2HG expression is markedly increased in hepatocellular carcinoma (HCC) and promotes the proliferation of cancer cells.15 However, the possible role of MIR4435-2HG in CC and the detailed mechanism of MIR4435-2HG on the occurrence and development of CC are still not fully revealed.
Recently, researchers found that microRNAs (miRNAs) serve as suppressors in human tumor genesis and many of them have also been applied as novel targets for cancer treatments.16–18 For instance, miR-584 can inhibit the viability, migration and invasion of CC in vitro.19 MiR-612 has been reported to inhibit the progression of CC.20 MiR-873 can suppress the viability of CC cells, thus retarding the development of CC.21 In addition, miR-128 has been reported to play an anti-cancer role in CC.22 A recent study has showed that up-regulation of miR-128 conspicuously inhibits the cell viability and epithelial-mesenchymal transition (EMT) procession in CC.22 However, the regulatory relationship between MIR4435-2HG and miR-128-3p in CC remains unclear.
In our study, the effects of MIR4435-2HG knockdown on the proliferation, migration and invasion of CC cells as well as the regulatory mechanism between MIR4435-2HG and miR-128-3p/Musashi 2 (MSI2) were investigated with a goal of providing a new treatment strategy for CC.
Patients and Methods
Tissues Collection
Totally, 59 CC patients (detailed data in Table 1) were selected in our hospital from 2016 to 2018. The inclusion criteria were as follows: 1. The patients were diagnosed as CC by pathological examinations. 2. None of the patients had received treatments for CC before the operation. 3. The clinical data of CC were complete. The patients according with the above conditions were chosen in this study and eventually the sample sizes were 59. The patients’ tumor and adjacent normal tissues were collected by surgery. Each patient in this study obtained the written informed consent. The protocols of this study were reviewed and approved by ethical committee of the Affiliated Hospital of Qingdao University.
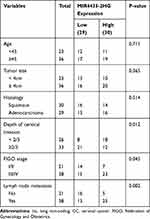 |
Table 1 Correlations Between lncRNA MIR4435-2HG Expression and Clinicopathological Characteristics in CC |
Cell Grouping and Transfection
The human CC cell lines (siha/hela) and the normal cervical cell line (HCerEpiC) were procured from Honsun Biotech, Ltd (Shanghai, China). The cells were grown in dulbecco’s modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2. siRNA-negative control (si-NC) and siRNA-MIR4435-2HG-1/-2 (si-MIR4435-2HG-1/-2) were procured from Sangon Biotech, Inc (Shanghai, China). Overexpression-MSI2 (pcDNA-MSI2) and its negative control (pcDNA-NC), miR-128-3p inhibitor, miR-128-3p mimics and their negative control (miR-NC) were all procured from Ribo Biotech, Ltd (Guangzhou, China). The aforementioned agents were transfected into the cells using Lipofectamine RNAiMAX kit (Invitrogen, Carlsbad, CA, USA) for 48 h. Following the transfection, the cells were harvested to perform the following experiments.
Bioinformatical Analysis
Based on The Cancer Genome Atlas (TCGA) database, MIR4435-2HG expression in both cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) and normal tissues was analyzed. Starbase software (http://starbase.sysu.edu.cn/) was used to predict the potential binding site between miR-128-3p and MIR4435-2HG, whereas Targetscan software (http://www.targetscan.org) was utilized to predict the potential binding site between miR-128-3p and MSI2.
Quantitative Reverse-Transcription PCR (qRT-PCR)
Following the manufacturer’s instructions, total RNA was extracted using TRIzol kit (Invitrogen, Inc.). The GoScript reverse transcription system (Promega, Madison, WI, USA) was used to reverse transcribe the extracted RNA into cDNA. Then the cDNA was subjected to qRT-PCR analysis. The thermocycling conditions were as follows: 94°C for 5 min, followed by 40 cycles at 94°C for 10 sec, 60°C for 40 sec and 72°C for 1 min. U6 or GADPH was used as the internal reference standards. Gene expression was quantified using the 2−ΔΔCt method.
Cell Viability Assay
The viability of CC cells was detected by 3-(4, 5-Dimethyl-2-Thiazolyl)-2, 5-Diphenyl-2-H-Tetrazolium Bromide (MTT) assay. The cells were seeded into a 96-well plate with 2 × 105 cells per well. Subsequently, the cells were incubated for 24, 48, 72 and 96 h. Then 20 µL MTT (Merck KGaA, Darmstadt, Germany) was added to each well at different time points. After 2h of incubation at 37°C, cell viability (optical density at 450 nm [OD450]) was analyzed using a Multiskan Spectrum microplate reader (Thermo Fisher Scientific).
Wound Healing Assay
Cells were seeded into 6-well plates and grown until 100% confluence in DMEM. Then the wounds on cell monolayer layers were created using a pipette tip. After that, the cells were incubated for 24 h in serum-free medium. Then, the cells were washed three times with phosphate-buffered saline (PBS) to wash away the floating cells. At 0 h and 24 h, the measurement and imaging of wound closure were measured by AxioVision v4.7 software (Carl Zeiss Meditec, Dublin, CA, USA) under a microscope (magnification, x 200).
Transwell Invasion Assay
Transwell chamber (Becton, Dickinson and Company, FL, NJ, USA) with Matrigel coating was used to detect the cell invasion. The upper chamber was seeded at a density of 2 × 105 cells per well in serum-free medium. DMEM in the lower chamber containing 10% FBS was as chemoattractant. After incubation for 3 h at 37°C with 5% CO2, cells in the lower chamber were stained with 0.5% crystal violet (Merck KGaA, Darmstadt, Germany) for 15 min at room temperature. A light microscope (magnification, x 400) was used to count the stained cells in five randomly-selected fields.
Dual-Luciferase Reporter (DLR) Assay
The wild-type (WT) vector was established by cloning 3ʹUTR sequences containing the binding site into pGL3 vector. The mutant-type (MUT) vector was constructed using the Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific). Wild-type/mutant-type vector and miR-128-3p mimics/miR-NC were co-transfected into the cells at 37°C. After 48 h of culture, a dual-luciferase reporter assay system (Promega) was used to detect the luciferase activity.
Western Blot Analysis
Radio-Immunoprecipitation Assay (RIPA) buffer containing protease inhibitors was used to extract proteins from cells, and the protein concentrations were detected by the Bicinchoninic Acid (BCA) Protein Assay Kit (Abcam, Cambridge, MA, USA). Approximately 40 µg proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Then, the resolving proteins were transferred into polyvinylidene fluoride (PVDF) membrane. Membrane blocking was performed using 5% bovine serum albumin (BSA) at room temperature. Next, the membrane was incubated overnight at 4°C with primary antibodies against MSI2 (1:500; Abcam) and β-actin (1:500; Abcam). Then, tris-buffered saline Tween-20 (TBST) was used to wash the membranes for three times. Subsequently, at room temperature, the HRP-conjugated anti-mice IgG secondary antibody (1:3000; Santa Cruz, Waltham, MA, USA) was added to incubate for 1 h. β-actin was used as the internal reference. Imprinting was detected using an electrochemiluminescence (ECL) detection kit (Millipore). ImageJ software (NIH) was utilised to quantify the image.
Statistical Analysis
SPSS Statistics software (vision 20.0, USA) was used to perform statistical analyses. Data are presented as the means ± standard deviations. Student’s t-test was used to assess the differences between two groups. One-way ANOVA was used to evaluate the differences among multiple groups. After ANOVA analysis, pairwise comparisons were assessed using Tukey’s multiple comparisons test. Pearson’s correlation analysis was used to determine the correlations between the expression of MIR4435-2HG and miR-128-3p as well as MSI2 and miR-128-3p in CC tissues. P-value less than 0.05 indicated a statistically significant difference. All experiments were conducted in triplicate in at least three independent trials.
Results
LncRNA MIR4435-2HG is Highly Expressed in CC/CESC Tissues and CC Cells
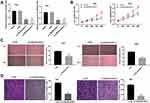
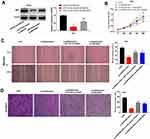
Based on the TCGA database, MIR4435-2HG expression in both CESC and normal tissues was analyzed. The results showed that MIR4435-2HG was significantly expressed in CESC tissues in comparison to that in normal tissues (Figure 1A, P < 0.05). Next, we detected the expression of MIR4435-2HG in CC and adjacent normal tissues from 59 patients to further verify the above results. qRT-PCR results demonstrated that MIR4435-2HG expression was higher in tumor tissues than the adjacent normal tissues (Figure 1B, P < 0.001). In line with this result, MIR4435-2HG expression was markedly increased in stage III/IV of CC compared to that in stage I/II (Figure 1C, P < 0.01). Additionally, we investigated the relationship between MIR4435-2HG expression and patients’ clinical parameters. The results showed that with the high expression and low expression of MIR4435-2HG exhibited significant differences in Federation of Gynecology and Obstetrics (FIGO) stage (P = 0.045) and lymph node metastasis (P = 0.002) (Table 1). Meanwhile, the expression of MIR4435-2HG in the CC cell lines was detected. qRT-PCR results revealed that MIR4435-2HG expression was significantly increased compared to that in HCerEpiC cells (Figure 1D, P < 0.01). Therefore, the data suggested that MIR4435-2HG might be an onco-lncRNA in CC.
LncRNA MIR4435-2HG Knockdown Inhibits the Proliferation, Migration and Invasion of CC Cells in vitro
Following transfection of si-miR4435-2HG-1/-2 into hela and siha cells, the efficiency of transfection was detected by qRT-PCR. The results showed that the expression of MIR4435-2HG was significantly decreased after transfection of si-miR4435-2HG-1 and si-miR4435-2HG-2 (Figure 2A, P < 0.01), which demonstrated that si-miR4435-2HG-1/-2 has been transfected into CC cells successfully. MTT assay showed that the viability of hela cells was dramatically reduced in the si-miR4435-2HG-1 group in comparison to the si-NC group, whilst similar result was obtained in siha cells (Figure 2B, P < 0.01). Wound healing assay uncovered that the wound healing rate of hela cells was reduced by knockdown of MIR-4435-2HG. Equally, the wound healing rate of siha cells was also reduced by the down-regulation of MIR-4435-2HG (Figure 2C, P < 0.01). Similarly, transwell invasion assay indicated that the number of invasion cells (hela and siha) was decreased by MIR4435-2HG knockdown (Figure 2D, P < 0.01). These results suggested that knockdown of MIR4435-2HG inhibited the proliferation, migration and invasion of CC cells in vitro.
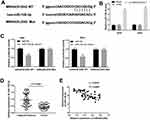
LncRNA MIR4435-2HG Targets miR-128-3p
Using Starbase software, the potential binding site between lncRNA MIR4435-2HG and miR-128-3p was depicted (Figure 3A). The expression of miR-128-3p was markedly up-regulated in the si-miR4435-2HG-1 group in contrast to the si-NC group in both two cell lines (Figure 3B, P < 0.01). DLR assay revealed that the luciferase activity in the MIR4435-2HG-WT/miR-128-3p mimics group was obviously decreased compared to the MIR4435-2HG-WT/miR-NC group (Figure 3C, P < 0.01). Moreover, miR-128-3p expression was down-regulated in tumor tissues in comparison to the adjacent normal tissues (Figure 3D, P < 0.001). Interestingly, a significant inverse correlation between MIR4435-2HG and miR-128-3p was exhibited in CC tissues (Figure 3E; P < 0.0001, r = −0.6331). These data implied that miR-128-3p was a direct target of MIR4435-2HG and was negatively modulated by MIR4435-2HG.
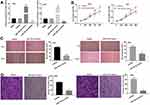
MiR-128-3p Suppresses the Proliferation, Migration and Invasion of CC Cells in vitro
To investigate the role of miR-128-3p on the biological functions of CC cells, the transfection efficiency of miR-128-3p was detected. qRT-PCR results uncovered that the expression of miR-128-3p was significantly increased by transfection of miR-128-3p mimics and was decreased by transfection of miR-128-3p inhibitor, suggesting that both miR-128-3p mimics and miR-128-3p inhibitor were successfully transfected into CC cells (Figure 4A, P < 0.01). MTT assay showed that up-regulation of miR-128-3p suppressed the cell viability in both two cell lines (Figure 4B, P < 0.01). Wound healing assay and transwell invasion assay respectively showed that the wound healing rate and number of invasion cells were both inhibited by miR-128-3p up-regulation (Figure 4C and D, P < 0.01). These data implied that up-regulation of miR-128-3p could inhibit the viability, migration ability and invasion ability of CC cells in vitro.
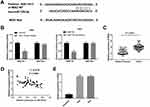
MiR-128-3p Targets MSI2
To explore whether miR-128-3p regulates MSI2, we predicted the potential binding site between miR-128-3p and MSI2 using Targetscan software (Figure 5A). Dual-luciferase reporter assay displayed that the luciferase activity was dramatically reduced with the overexpression of miR-128-3p in the MSI2-WT group compared to that in the MSI2-MUT group. The results uncovered that miR-128-3p targeted MSI2 (Figure 5B, P < 0.01). Furthermore, qRT-PCR results demonstrated that the expression of MSI2 was higher in tumor tissues (Figure 5C, P < 0.001) or CC cells (Figure 5E, P < 0.01) than those in adjacent normal tissues or HCerEpiC cells. At the same time, we found that miR-128-3p expression was negatively correlated with MSI2 (Figure 5D; P < 0.0001, r = −0.4775). In brief, the above results demonstrated that MSI2 was a target gene of miR-128-3p and was negatively modulated by miR-128-3p.
MIR4435-2HG Knockdown Inhibits the Proliferation, Migration and Invasion of CC Cells in vitro by Regulating the miR-128-3p/MSI2 Axis
Western blot analysis demonstrated that the protein level of MIS2 in hela cells was down-regulated in the miR-128-3p mimics + pcDNA-NC group in contrast to the miR-NC + pcDNA-NC group, whilst this situation was reversed in the miR-128-3p mimics + pcDNA-MSI2 group (Figure 6A, P < 0.01). MTT assay uncovered that the viability of hela cells was significantly inhibited in the si-miR4435-2HG-1 group in comparison to the si-NC group. Meanwhile, cell viability was partly promoted in both the si-miR4435-2HG-1 + pcDNA-MIS2 group (Figure 6B, P < 0.05) and the si-miR4435-2HG-1 + miR-128-3p inhibitor group (P < 0.05) in comparison to the si-miR4435-2HG-1 group. In a similar vein, wound healing assay and transwell invasion assay respectively showed that the wound healing rate and number of invasion cells were inhibited by MIR4435-2HG knockdown. However, both down-regulation of miR-128-3p and up-regulation of MSI2 reversed the inhibiting effects of MIR4435-2HG knockdown on the migration and invasion in hela cells (Figure 6C and D, P < 0.01). These results implied that knockdown of MIR4435-2HG could suppress the viability, migration ability and invasion ability of CC cells through regulating the miR-128-3p/MSI2 axis.
Discussion
CC is one of the most serious cancer in females and a leading cause of tumor-related death among women globally.1,23 LncRNAs have been reported to promote tumorigenesis in multiple cancers, such as cervical,24 colorectal14 and prostate cancer.25 A recent study has displayed that the expression of CAR10 in CC tissues and cells is dramatically increased, whereas up-regulation of CAR10 is closely correlated with lymph node metastasis.11 In another study, OIP5-AS1 expression is markedly up-regulated in CC cells, which is significantly associated with lymph node metastasis and tumor stage.9 Similarly, we found that MIR4435-2HG expression was dramatically increased in CC tissues and cells. Besides MIR4435-2HG expression exhibited significant correlations with FIGO stage and lymph node metastasis. Therefore, we thought that MIR4435-2HG might be a pathogenic factor in CC.
In the past decade, researchers have determined that lncRNAs act as important regulators in the proliferation, migration and invasion of CC cells.10,11,26 Song et al have disclosed that down-regulation of OIP5-AS1 inhibits viability and promotes apoptosis in CC cells.9 Zhu et al have disclosed that silencing of CDKN2B-AS1 suppresses metastasis and promotes apoptosis in CC cells.27 Liu et al have demonstrated that inhibition of lncRNA GHET1 decreases the viability and migration ability of CC cells.28 In the current study, we found that lncRNA MIR4435-2HG knockdown suppressed the viability, migration ability and invasion ability of CC cells. Similar to our results, a recent study has demonstrated that the proliferation, invasion and migration of CRC cells can be inhibited by MIR4435-2HG knockdown.14 However, the previous study only investigated the regulatory mechanism of MIR4435-2HG on CRC. Our results further confirmed that lncRNA MIR4435-2HG also accelerated CC progression.
Accumulating evidence indicates that miRNAs participate in the progression of tumors29 and serve as anti-tumor roles in several tumor types.30–32 Han et al have uncovered that miR-128 is down-regulated in gastric cancer (GC), and overexpression of miR-128 reduces the proliferation and induces the apoptosis in GC cells.33 Sun et al have found that miR-128 expression is obviously decreased in prostate cancer (PC) tissues and overexpression of miR-128 restrains the invasion of PC cells.34 In this study, we demonstrated that miR-128-3p was obviously decreased in CC tissues. The proliferation, migration, and invasion of CC cells could be inhibited by miR-128-3p overexpression. In line with our results, Wu et al have showed that miR-128 is down-regulated in CC cells, whereas up-regulation of miR-128 obviously suppresses the proliferation, migration, and invasion in CC in vitro.22 Moreover, we also proved miR-128-3p was a target of MIR4435-2HG. The results implied that miR-128-3p might be regulated by MIR4435-2HG to inhibit the development of CC.
The emerging evidence suggests that MSI2 is an important factor in the development of cancers.35 Wang et al have presented that MSI2 is significantly expressed in HCC tissues, thus promoting the migration and invasion of HCC cells.36 Kudinov et al have reported that overexpression of MSI2 accelerates metastasis in mouse non-small cell lung cancer (NSCLC) cells and human NSCLC tumors.37 In current study, we demonstrated that MSI2 expression in CC tissues and cells was significantly increased. In line with this result, Liu et al have showed that MSI2 expression is dramatically up-regulated in human CC tissues.35 Meanwhile, MSI2 was proved to be a target gene of miR-128-3p and there was a markedly inverse correlation between miR-128 and MSI2 in CC tissues. The results uncovered that MSI2 was negatively regulated by miR-128-3p. Simultaneously, we revealed that both down-regulation of miR-128-3p and up-regulation of MSI2 reversed the suppressive effects of MIR4435-2HG knockdown on the proliferation, migration, and invasion of hela cells. The results suggested that MIR4435-2HG knockdown inhibited the proliferation, migration, and invasion of CC cells through regulating the miR-128/MSI2 axis.
Conclusions
To be concluded, the current study uncovered that knockdown of MIR4435-2HG could suppress the proliferation, migration and invasion of CC cells through regulating the miR-128/MSI2 axis in vitro. However, this study did not conduct validation experiments in vivo. This may be a limitation of the present study. Further studies to elucidate these issues will be performed in the future. Even so, we also hope these findings will provide a new strategy for treating CC.
Ethics Approval and Consent to Participate
This study was approved by the ethics committee of The Affiliated Hospital of Qingdao University. Written informed consent was obtained from all subjects.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2000;370(9590):890–907. doi:10.1016/S0140-6736(07)61416-0
2. Jin X, Chen X, Hu Y, Ying F, Zhu H. LncRNA-TCONS_00026907 is involved in the progression and prognosis of cervical cancer through inhibiting miR-143-5p. Cancer Med. 2017;6(6):1409–1423. doi:10.1002/cam4.1084
3. Jemal A, Bray F, Center MM, Ferlay J, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;6(2):169–190.
4. Franco EL, Duarte-Franco E, Ferenczy A. Cervical cancer: epidemiology, prevention, and the role of human papillomavirus infection. Cmaj. 2001;164(7):1017–1025.
5. Vaccarella S, Lortettieulent J, Plummer M, Franceschi S, Bray F. Worldwide trends in cervical cancer incidence: impact of screening against changes in disease risk factors. Eur J Cancer. 2013;49(15):3262–3273. doi:10.1016/j.ejca.2013.04.024
6. Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis. 2018;9(6):706. doi:10.1038/s41419-018-0724-5
7. Tao F, Xinxin T, Shanming R, Minhe S, Zhiqian Z. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. FASEB J. 2018;32(11):6330–6343.
8. Zhu J, Shi H, Liu H, Wang X, Li F. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017;8(39):65253–65264. doi:10.18632/oncotarget.18224
9. Song L, Wang L, Pan X, Yang C. lncRNA OIP5-AS1 targets ROCK1 to promote cell proliferation and inhibit cell apoptosis through a mechanism involving miR-143-3p in cervical cancer. Braz J Med Biol Res. 2020;53(1):e8883. doi:10.1590/1414-431x20198883
10. Ma Z, Gu G, Pan W, Chen X. LncRNA PCAT6 accelerates the progression and chemoresistance of cervical cancer through up-regulating ZEB1 by sponging miR-543. Onco Targets Ther. 2020;13:1159–1170. doi:10.2147/OTT.S232354
11. Hu T, Zhang Q, Gao L. LncRNA CAR10 upregulates PDPK1 to promote cervical cancer development by sponging miR-125b-5p. Biomed Res Int. 2020;2020:4351671. doi:10.1155/2020/4351671
12. Qian H, Chen L, Huang J, et al. The lncRNA MIR4435-2HG promotes lung cancer progression by activating beta-catenin signalling. J Mol Med (Berl). 2018;96(8):753–764. doi:10.1007/s00109-018-1654-5
13. Ouyang W, Ren L, Liu G, et al. LncRNA MIR4435-2HG predicts poor prognosis in patients with colorectal cancer. Peerj. 2019;7:e6683. doi:10.7717/peerj.6683
14. Dong X, Yang Z, Yang H, Li D, Qiu X. Long non-coding RNA MIR4435-2HG promotes colorectal cancer proliferation and metastasis through miR-206/YAP1 axis. Front Oncol. 2020;10:160. doi:10.3389/fonc.2020.00160
15. Kong Q, Liang C, Jin Y, Pan Y, Tong D, Zhou J. The lncRNA MIR4435-2HG is upregulated in hepatocellular carcinoma and promotes cancer cell proliferation by upregulating miRNA-487a. Cell Mol Biol Lett. 2019;24:26. doi:10.1186/s11658-019-0148-y
16. Shah A, Leidinger P, Blin N, Meese E. miRNA: small molecules as potential novel biomarkers in cancer. Curr Med Chem. 2010;17(36):4427–4432. doi:10.2174/092986710794182980
17. Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104(2):235–240. doi:10.1038/sj.bjc.6606010
18. Tazawa H, Kagawa S, Fujiwara T. MicroRNAs as potential target gene in cancer gene therapy of gastrointestinal tumors. Expert Opin Biol Ther. 2011;11(2):145–155. doi:10.1517/14712598.2011.542749
19. Wang T, Feng J, Zhang A. miR-584 inhibits cell proliferation, migration and invasion in vitro and enhances the sensitivity to cisplatin in human cervical cancer by negatively targeting GLI1. Exp Ther Med. 2020;19(3):2059–2066. doi:10.3892/etm.2020.8449
20. Jin Y, Zhou X, Yao X, Zhang Z, Cui M, Lin Y. MicroRNA-612 inhibits cervical cancer progression by targeting NOB1. J Cell Mol Med. 2020;24(5):3149–3156. doi:10.1111/jcmm.14985
21. Feng J, Wang T. MicroRNA-873 serves a critical role in human cervical cancer proliferation and metastasis via regulating glioma-associated oncogene homolog 1. Exp Ther Med. 2020;19(2):1243–1250. doi:10.3892/etm.2019.8348
22. Wu W, Guo L, Liang Z, Liu Y, Yao Z. Lnc-SNHG16/miR-128 axis modulates malignant phenotype through WNT/beta-catenin pathway in cervical cancer cells. J Cancer. 2020;11(8):2201–2212. doi:10.7150/jca.40319
23. Chen X, Xiong D, Yang H, et al. Long noncoding RNA OPA-interacting protein 5 antisense transcript 1 upregulated SMAD3 expression to contribute to metastasis of cervical cancer by sponging miR-143-3p. J Cell Physiol. 2019;234(4):5264–5275. doi:10.1002/jcp.27336
24. Fan Y, Sheng W, Meng Y, Cao Y, Li R. LncRNA PTENP1 inhibits cervical cancer progression by suppressing miR-106b. Artif Cells Nanomed Biotechnol. 2020;48(1):393–407. doi:10.1080/21691401.2019.1709852
25. Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65(6):1140–1151. doi:10.1016/j.eururo.2013.12.003
26. Chen WJ, Xiong L, Yang L, et al. Long non-coding RNA LINC01783 promotes the progression of cervical cancer by sponging miR-199b-5p to mediate GBP1 expression. Cancer Manag Res. 2020;12:363–373. doi:10.2147/CMAR.S230171
27. Zhu L, Zhang Q, Li S, Jiang S, Cui J, Dang G. Interference of the long noncoding RNA CDKN2B-AS1 upregulates miR-181a-5p/TGFbetaI axis to restrain the metastasis and promote apoptosis and senescence of cervical cancer cells. Cancer Med. 2019;8(4):1721–1730. doi:10.1002/cam4.2040
28. Liu Z, Luo S, Wu M, Huang C, Shi H, Song X. LncRNA GHET1 promotes cervical cancer progression through regulating AKT/mTOR and Wnt/beta-catenin signaling pathways. Biosci Rep. 2020;40(1):BSR20191265. doi:10.1042/BSR20191265
29. Nouraee N, Khazaei S, Vasei M, Razavipour SF, Mowla SJ. MicroRNAs contribution in tumor microenvironment of esophageal cancer. Cancer Biomarkers. 2016;16(3):367. doi:10.3233/CBM-160575
30. Zhang Y, Wang Y, Wang J. MicroRNA-584 inhibits cell proliferation and invasion in non-small cell lung cancer by directly targeting MTDH. Exp Ther Med. 2018;15(2):2203–2211.
31. Xue H, Guo X, Han X, Yan S, Li G. MicroRNA-584-3p, a novel tumor suppressor and prognostic marker, reduces the migration and invasion of human glioma cells by targeting hypoxia-induced ROCK1. Oncotarget. 2015;7(4):4785–4805. doi:10.18632/oncotarget.6735
32. Wang XP, Deng XL, Li LY. MicroRNA-584 functions as a tumor suppressor and targets PTTG1IP in glioma. Int J Clin Exp Pathol. 2014;7(12):8573.
33. Han L, Xiong L, Wang C, Shi Y, Song Q, Sun G. MicroRNA-128 contributes to the progression of gastric carcinoma through GAREM-mediated MAPK signaling activation. Biochem Biophys Res Commun. 2018;504(1):295–301. doi:10.1016/j.bbrc.2018.08.177
34. Sun X, Li Y, Jie Y, Hong P, Luo P, Jie Z. miR-128 modulates chemosensitivity and invasion of prostate cancer cells through targeting ZEB1. Jpn J Clin Oncol. 2015;45(5):474–482. doi:10.1093/jjco/hyv027
35. Liu Y, Fan Y, Wang X, Huang Z, Shi K, Zhou B. Musashi-2 is a prognostic marker for the survival of patients with cervical cancer. Oncol Lett. 2018;15(4):5425–5432. doi:10.3892/ol.2018.8077
36. Wang MH, Qin SY, Zhang SG, Li GX, Peng ZH. Musashi-2 promotes hepatitis B virus related hepatocellular carcinoma progression via the Wnt/β-catenin pathway. Am J Cancer Res. 2015;5(3):1089–1100.
37. Kudinov AE, Deneka A, Nikonova AS, Beck TN, Boumber Y. Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc Natl Acad Sci. 2016;113(25):6955–6960. doi:10.1073/pnas.1513616113
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.