Back to Journals » OncoTargets and Therapy » Volume 13
Knockdown of Long Non-Coding RNA LOC100132707 Inhibits the Migration of Uveal Melanoma Cells via Silencing JAK2
Authors Qi Y, Yao R , Zhang W , Cui Q , Zhang F
Received 9 July 2020
Accepted for publication 29 October 2020
Published 17 December 2020 Volume 2020:13 Pages 12955—12964
DOI https://doi.org/10.2147/OTT.S266596
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Ying Qi, Renjie Yao, Wenjing Zhang, Qingqing Cui, Fengyan Zhang
Department of Ophthalmology, The First Affiliated Hospital of Zhengzhou University, The Laboratory for Ophthalmology and Vision Science, Henan Eye Hospital, Zhengzhou 450052, Henan, People’s Republic of China
Correspondence: Ying Qi; Fengyan Zhang
Department of Ophthalmology, The First Affiliated Hospital of Zhengzhou University, The Laboratory for Ophthalmology and Vision Science, Henan Eye Hospital, Zhengzhou 450052, Henan, People’s Republic of China
Tel +86-371-3758296
Email [email protected]; [email protected]
Background/Objective: Although lots of long non-coding RNAs (lncRNAs) have been demonstrated to be involved in carcinogenesis, the functions of numerous of lncRNAs remain unknown. Bioinformatics online database showed that lncRNA LOC100132707 was highly expressed in metastatic melanoma tissues, and its expression predicted a lower overall survival rate in melanoma patients. However, LOC100132707 function in uveal melanoma (UM) progression still remains unclear. In the present study, we aimed to elucidate the role and molecular mechanisms underlying LOC100132707 in UM.
Methods: RT-PCR was used to detect the levels of LOC100132707 in UM cells. Cell migration, invasion and tumorigenesis were tested by using the transwell chamber assay and in vivo assay.
Results: LOC100132707 expression in metastatic UM cell line MM28 was significantly higher than that of the non-metastatic UM cell lines, MP38, MP46 and MP65, as well as the expressions of LOC100132707-related genes, including XRN1, PARP14, JAK2, DDX60, BUB1 and SAMD9L. LOC100132707 downregulation significantly repressed cell migration and invasion abilities, whereas overexpressing JAK2 rescued these effects. Consistently, upregulation of LOC100132707 induced significant increases in cell migration and invasion abilities via upregulating JAK2. In addition, silencing of LOC100132707 significantly repressed the in vivo tumor formation ability in UM cells.
Conclusion: This study reveals that silence of LOC100132707 represses the migration of UM via downregulating JAK2. The LOC100132707/JAK2 axis might serve as a potent target for the prevention and treatment of UM metastasis.
Keywords: long non-coding RNA, lncRNA, LOC100132707, metastasis, uveal melanoma, UM, JAK2
Introduction
Uveal melanoma (UM) is the most common intraocular malignancy in adults, accounting for ~5% among all kinds of melanomas.1,2 It’s estimated that approximately seven cases from per million people suffer from this disease every year,1 bringing great threats to human visual acuity and even life.3,4 Despite effective command of the primary tumor, nearly 50% patients develop metastatic disease to the liver within 15 years.5 The 5-year survival rate for patients with UM has not been obviously improved over the past 40 years as the high metastasis rates.5 Therefore, it’s urgent to further reveal the molecular mechanism underlying the metastasis of UM.
Long non-coding RNA (lncRNA) is a type of non-coding RNA molecular with more than 200 nucleosides in length. As the development of sequencing technology, more and more lncRNAs have been found and identified to play important roles in the regulation of cancer cell proliferation, apoptosis, metastasis and drug resistance.6–8 For instance, lncRNA loc285194 was identified to be a p53-regulated tumor suppressor, that partially inhibited tumor growth via inhibiting microRNA-211.9 Noticeably, lncRNAs have been also demonstrated to be closely involved in the development and progression of melanoma.10 Schmidt et al11 showed that lncRNA SLNCR1 expression was elevated in cutaneous melanoma and promoted melanoma invasion. Evidence has demonstrated that lncRNAs can serve as competitive RNAs (ceRNA) which abrogated the inhibitory effect of miRNA on their target mRNAs, leading to the development of multiple kinds of diseases including cancers.12 For example, lncRNA LINC01234 acted as a ceRNA in osteosarcoma, and regulated CBFB expression via mutagenizing miR-204-5p.13 Liang et al14 reported that lncRNA ZFAS1 promoted tumorigenesis through regulating the expression of miR-150-5p/RAB9A axis in melanoma.
In the present study, we used the Cancer RNA-Seq Nexus Database (https://omictools.com/crn-tool) to find the different expression lncRNAs in the metastatic melanoma. The results showed that lncRNA LOC100132707 (Genecards ID: PAXIP1-AS2) is the most different expressed gene among primary melanoma and metastatic melanoma. However, its role in the progression of UM remains unknown, as well as the underlying mechanisms. Therefore, we aimed to reveal the role and mechanism underlying lncRNA LOC100132707 in the occurrence and development of UM.
Materials and Methods
Ethic Statement
The current study obtained the approval of the Ethic Committee of Henan Eye Hospital (Henan province, China).
Bioinformatics Analysis
Differently expressed lncRNAs among primary melanoma and metastatic melanoma were assessed by Cancer RNA-Seq Nexus Database (https://omictools.com/crn-tool). The expression patterns of lncRNA LOC100132707 were evaluated using The Cancer Genome Atlas (TCGA) data portal. GEPIA database (http://gepia.cancer-pku.cn/) was used to find genes which are related to lncRNA LOC100132707 in melanoma.
Cell Lines and Culture
Four UM cell lines, MP38, MP46, MP65 and MM28 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and were grown in RPMI-1640 medium, supplemented with 20% fetal bovine serum (FBS, Gibco, MA, USA).
Cell Transfection
The short hairpin RNAs (shRNAs) used to silence LOC100132707 (sh-LOC100132707) in UM cells and its negative control vector (sh-NC), and the LOC100132707 overexpressing vector (OE-LOC100132707) and OE-NC were synthesized by GenePharma (Shanghai, China). Vector used to overexpress Janus kinase 2 (JAK2) (OE-JAK2; No. SC316317) and sh-JAK2 (No. TL320395) were purchased from OriGene (Beijing, China). sh-LOC100132707, sh-JAK2 and sh-NC vectors were induced into cells with the help of 7 µg/mL polybrene. OE-JAK2, OE-LOC100132707 and OE-NC were transfected into cells using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. In addition, cells were incubated with polybrene (4 µg/mL) for 14 days to induce stable cell lines which were used in the in vivo assay.
Western Blotting
Cells or tissues were homogenised with lysis buffer (Solarbio, Beijing, China) with protease inhibitor cocktail (Sigma Aldrich, Santa Clara, CA, USA) according to the manufacturer’s instructions. Following protein concentration being determined with the BCA (bicinchoninic acid) protein assay kit (Thermo Scientific, MA, USA), protein samples were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA). The membranes were then incubated with the primary antibodies targeting JAK2 (No. #3230, Cell Signaling Technology, MA, USA), MMP2 (No. ab97779, Abcam, Cambridge, MA, USA), MMP9 (No. ab119906, Abcam) or GAPDH (No. #5174, Cell Signaling Technology) protein at 4 °C overnight, followed by incubation with the second antibody for 1 h at room temperature. The blots were detected with the enhanced chemiluminescence detection kit (Millipore). The specific protein bands were analyzed using the Image J software with the density values of GAPDH as an internal reference.
Real-Time PCR (RT-PCR)
Total RNA was extracted from cells and tissues using RNAsimple total RNA extract kit (CWBIO, Jiangsu, China) according to the manufacturer’s instructions. After quantification with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, Del, USA), a total of 1 µg RNA was reversely transcribed into cDNA with the help of a First Strand cDNA Synthesis Kit (CWBIO). RT-PCR was carried out on an ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) with an SYBR Green (CWBIO). GAPDH was used as an internal reference to normalize mRNA expression. Primer sequences are listed in Table 1.
 |
Table 1 Primer Sequences Used in RT-PCR |
Transwell Chamber Assay
Cell migration was detected by using Transwell chambers (BD Bioscience). A total of 1×105 UM cells infected with different lentivirus vectors were inoculated into the top chamber and incubated at 37 °C, 5% CO2 for 24 hours. Next, cells on the bottom of the filter were fixed in 4% paraformaldehyde and stained with 1% crystal violet (Solarbio). Nest, the migrated cells were counted under a microscope. Invasion assay was carried out as same as the migration assay with Matrigel pre-coated chambers and the cells were allowed to invade for 48 hours. Each assay was performed in triplicate with five random images per filter.
In vivo Assay
The animal studies were approved by the Ethic Committee of Henan Eye Hospital. Briefly, MM28 cells (5× 106 cells in 200 μL PBS/mouse) with sh-NC or sh-LOC100132707 stable transfection were subcutaneously injected into the flanks of male (6-week-old) NOD/SCID mice (Vitalriver, Beijing, China). Animals were euthanized and the xenografts were dissected and weighed following 28 days of injection.
Statistical Analysis
All the quantitative data are presented as means ± standard deviation (SD). The statistical significance of differences was determined by using Student’s two-tailed t test in two groups, and one-way ANOVA followed with Tukey’s test in multiple groups. A p value <0.05 was considered statistically significant.
Results
LOC100132707 is Overexpressed in Melanoma Tissues and UM Cells
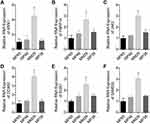
First, we used bioinformatics methods to search the differently expressed lncRNAs between primary melanoma and metastatic melanoma. The results showed that several lncRNAs were upregulated in the metastatic melanoma, such as LOC100132707, LOC339524, FLJ41200, LOC100505812, LOC153684, OIP5-AS1, LOC100506585, etc. (Figure 1A). Among them, LOC100132707 expression patterns showed the biggest difference; hence, we focused on LOC100132707 and aimed to reveal its roles in UM progression. The high expression pattern of LOC100132707 in the metastatic melanoma was also confirmed by the data uploaded from the TCGA database (Figure 1B). Moreover, the high expression profile of LOC100132707 predicted a shorter overall survival in patients with melanoma (Figure 1C). Similarly to LOC100132707 expression pattern in metastatic melanoma and primary melanoma samples, its expression was significantly higher in the metastatic human UM cell strain MM28 than that in the non-metastatic UM cell lines, such as MP38, MP46 and MP65 (Figure 1D). These results suggested that the high expression of LOC100132707 might be involved in the metastasis of UM.
LOC100132707-Related Genes Associates with UM Metastasis
Next, we analyzed the expression patterns of LOC100132707-related genes in the metastatic human UM cell strain MM28 and the non-metastatic UM cell lines, such as MP38, MP46 and MP65. The mRNA expressions of XRN1, PARP14, JAK2, DDX60, BUB1 and SAMD9L were significantly higher in MM28 cells than that in MP38, MP46 and MP65 cells (Figure 2A–F). These results further suggested that LOC100132707 might be involved in UM metastasis via regulating its related genes.
LOC100132707 Expression Positively Associates with XRN1, PARP14, JAK2, DDX60, BUB1 and SAMD9L
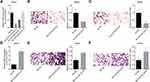
We then using Pearson correlation test to evaluate the correlations between LOC100132707 expression and XRN1, PARP14, JAK2, DDX60, BUB1 and SAMD9L expressions. The results showed that LOC100132707 expression showed positive association with XRN1, PARP14, JAK2, DDX60, BUB1 and SAMD9L levels (Figure 3A–F).
LOC100132707 Promotes the Migration and Invasion of UM Cells
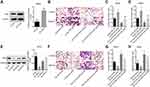
Next, we performed gain- and loss-of-function assays in MP65 and MM28 cells to explore LOC100132707 effects on cell migration and invasion, respectively. The expression of LOC100132707 was significantly reduced when MM28 cells were transfected with sh-LOC100132707-1 and sh- LOC100132707-2 (Figure 4A), while LOC100132707 expression was increased when MP65 cells were transfected with OE-LOC100132707 (Figure 4D). Knockdown of LOC100132707 significantly repressed cell invasion and migration in MM28 cells (Figure 4B and C), whereas overexpression of LOC100132707 induced significant promotions in cell invasion and migration abilities in MP65 cells (Figure 4E and F). These results demonstrated that LOC100132707 served as a promoter in UM metastasis.
LOC100132707 Promotes Cell Migration and Invasion via Upregulating JAK2 Expression in UM Cells
To reveal the mechanism underlying LOC100132707 in promoting the metastasis of UM cells, we then carried out the rescued experiments. OE-JAK2 transfection induced a 3-fold increase in JAK2 protein expression level in MM28 cells (Figure 5A). Transwell chamber assays showed that JAK2 overexpression significantly rescued sh-LOC100132707 role in inhibiting MM28 cell migration and invasion (Figure 5B–D). In addition, JAK2 was silenced and LOC100132707 was upregulated in MP65 cells to revel the role of LOC100132707/JAK2 axis in cell invasion and migration. Western blotting result showed that sh-JAK2-1 and sh-JAK2-2 significantly reduced JAK2 protein expression level in MP65 cells (Figure 5E). sh-JAK2-1 was chosen for the following study. The transwell chambers results showed that knockdown of JAK2 obviously blunted OE-LOC100132707 effects on the promotions of cell invasion and migration in MP65 cells (Figure 5F–H). These findings suggested that LOC100132707 facilitated UM cell migration and invasion via upregulating JAK2.
Knockdown of LOC100132707 Significantly Weakens the Tumorigenesis of UM Cells
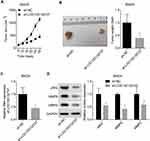
Additionally, we explored LOC100132707 role in the progression of UM in vivo. Compared with the control group, downregulation of LOC100132707 significantly decreased the in vivo tumor formation ability of MM28 cells (Figure 6A–C), with decreased expression levels of JAK2, and the metastasis markers MMP2 and MMP9 (Figure 6D). These results confirmed the inhibitory role of LOC100132707 in UM progression.
Discussion
According to the data of global transcriptional analysis, a wide range of lncRNAs are generated from the transcription of human genome.15 Increasing evidence has indicated that lncRNAs play important roles in the malignant transformation of multiple kinds of tumors including UM. For instance, Lu et al16 reported that the expression of lncRNA RHPN1 antisense RNA 1 was significantly increased in UM cell lines, and knockdown of it obviously inhibited UM cell proliferation and migration in vitro and in vivo. Similarly, lncRNA HOXA11-AS expression was increased in UM, leading to cancer progression.17 LncRNA PVT1 was also reported to be highly expressed in UM tissues, and downregulation induced a significant reduction in cell proliferation and an increase in cell apoptosis.18 These aforementioned lncRNAs are overexpressed in UM samples and serve as oncogenes. However, some lncRNAs exert opposite roles. Ding et al19 revealed that lncRNA PAUPAR expressed in UM tissues and cells at low levels; ectopic expression of it repressed the tumourigenesis of UM in vitro and in vivo. Xing et al20 demonstrated that lncRNA CASC15-New-Transcript 1 (CANT1) significantly reduced tumor metastatic ability and tumor formation in cultured cells and tumor-harboring animals. These above studies illustrate that lncRNAs are of crucial in UM progression and might serve as the potent targets for UM treatment.
As almost 50% of patients with UM develop metastatic disease to the liver,21 we mainly aimed to reveal the molecular mechanisms underlying UM metastasis. In the present study, we explored the expression pattern and function of lncRNA LOC100132707 (Genecards ID: PAXIP1-AS2) in UM for first time. LOC100132707 located at 7q36.2 is a ~1.71 kb RNA transcribed from the chromosome 7. From the NCBI database (https://www.ncbi.nlm.nih.gov/gene/100132707), we find that LOC100132707 is widely expressed in human tissues, such as colon, endometrium, esophagus, spleen, ovary, etc. It has been demonstrated that LOC100132707 level was increased in glioblastoma samples when compared to normal brain tissues, which was associated with the malignancy grades of glioma.22 In the present study, we found that LOC100132707 expression was apparently increased in the metastatic melanoma using bioinformatics analysis, as well as its expression level in the metastatic UM cell line MM28, suggesting that the deregulation of LOC100132707 might be involved in the metastasis of UM. To determine its function in UM metastasis, we then carried out gain- and loss-of function experiments in UM cells. The results showed that overexpression of LOC100132707 significantly promoted UM cell migration and metastasis, while knockdown of LOC100132707 inhibited cell migration and invasion. Further experiments also showed that knockdown of LOC100132707 also induced asignificant repression in the tumorigenesis in vivo, together with apparent decreases in the expression levels of metastasis markers (MMP2 and MMP9). These results confirmed that LOC100132707 serves as an accelerator in UM cell metastasis.
In mechanism, we found that the expression levels of XRN1, PARP14, JAK2, DDX60, BUB1 and SAMD9L showed positive correlation to LOC100132707 expression pattern in melanoma, suggesting that they might be implicated in LOC100132707-mediated metastasis in UM. JAK2 is a principal member of the JAK2/STAT3 (signal transducer and activator of transcription) signaling, which is identified to be frequently overactivated and closely involved in the occurrence of many human cancers, including melanoma.23–25 Activated JAK2 triggers STAT3 activation and nucleus translocation by inducing tyrosine phosphorylation, which then leads to transcription and translation of downstream genes.26 Therefore, targeting JAK2 signaling is a potential anticancer strategy.27 Herein, we explored JAK2 function in LOC100132707-mediated UM progression. We observed that JAK2 downregulation weakened LOC100132707 roles in promoting UM cell migration and invasion, indicating that JAK2 played an indispensable role in LOC100132707-mediated enhancements in UM cell migration and invasion.
However, the present study still has several limitations. One is that we did not explore the expression pattern of LOC100132707 in primary UM tissues and metastasis tissues. The other is that we did not explore the effect of JAK2/STAT3 signaling on sh-LOC100132707-mediated inhibition in tumor formation in vivo. We intend to solve the above problems in our further study.
In summary, this study reveals that silence of LOC100132707 can repress the migration and invasion of UM via downregulating JAK2. Our results indicate that LOC100132707/JAK2 axis might serve as a potent target for the prevention and treatment of UM metastasis.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Miyamoto C, Balazsi M, Bakalian S, Fernandes BF, Burnier MN
2. Field MG, Durante MA, Anbunathan H, et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S. Nat Commun. 2018;9(1):116. doi:10.1038/s41467-017-02428-w
3. Bakalian S, Marshall JC, Logan P, et al. Molecular pathways mediating liver metastasis in patients with uveal melanoma. Review. Clin Cancer Res. 2008;14(4):951–956. doi:10.1158/1078-0432.CCR-06-2630
4. Moore AR, Ran L, Guan Y, et al. GNA11 Q209L mouse model reveals RasGRP3 as an essential signaling node in uveal melanoma. Research Support, U.S. Gov’t, Non-P.H.S. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. Cell Rep. 2018;22(9):2455–2468. doi:10.1016/j.celrep.2018.01.081
5. Yang J, Manson DK, Marr BP, Carvajal RD. Treatment of uveal melanoma: where are we now? Review. Ther Adv Med Oncol. 2018;10:1758834018757175. doi:10.1177/1758834018757175
6. Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Review. Cell Oncol (Dordr). 2018;41(6):585–603. doi:10.1007/s13402-018-0406-4
7. Huarte M. The emerging role of lncRNAs in cancer. Research Support, Non-U.S. Gov’t Review. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
8. Chen LL. Linking long noncoding RNA localization and function. Review Research Support, Non-U.S. Gov’t. Trends Biochem Sci. 2016;41(9):761–772. doi:10.1016/j.tibs.2016.07.003
9. Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Research Support, N.I.H., Extramural. Nucleic Acids Res. 2013;41(9):4976–4987. doi:10.1093/nar/gkt182
10. Leucci E, Coe EA, Marine JC, Vance KW. The emerging role of long non-coding RNAs in cutaneous melanoma. Review. Pigment Cell Melanoma Res. 2016;29(6):619–626. doi:10.1111/pcmr.12537
11. Schmidt K, Joyce CE, Buquicchio F, et al. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep. 2016;15(9):2025–2037. doi:10.1016/j.celrep.2016.04.018
12. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Review. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
13. Chen X, Chen Z, Yu S, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018;24(8):2002–2014. doi:10.1158/1078-0432.CCR-17-2376
14. Liang L, Zhang Z, Qin X, et al. Long noncoding RNA ZFAS1 promotes tumorigenesis through regulation of miR-150-5p/RAB9A in melanoma. Melanoma Res. 2019;29(6):569–581. doi:10.1097/CMR.0000000000000595
15. Lee JT. Epigenetic regulation by long noncoding RNAs. Research support, Non-U.S. Gov’t Review. Science. 2012;338(6113):1435–1439. doi:10.1126/science.1231776
16. Lu L, Yu X, Zhang L, et al. The long non-coding RNA RHPN1-AS1 promotes uveal melanoma progression. Int J Mol Sci. 2017;18(1):Jan. doi:10.3390/ijms18010226
17. Lu Q, Zhao N, Zha G, Wang H, Tong Q, Xin S. LncRNA HOXA11-AS exerts oncogenic functions by repressing p21 and miR-124 in uveal melanoma. DNA Cell Biol. 2017;36(10):837–844. doi:10.1089/dna.2017.3808
18. Huang XM, Shi SS, Jian TM, Tang DR, Wu T, Sun FY. LncRNA PVT1 knockdown affects proliferation and apoptosis of uveal melanoma cells by inhibiting EZH2. Eur Rev Med Pharmacol Sci. 2019;23(7):2880–2887. doi:10.26355/eurrev_201904_17566
19. Ding X, Wang X, Lin M, et al. PAUPAR lncRNA suppresses tumourigenesis by H3K4 demethylation in uveal melanoma. Letter Research Support, Non-U.S. Gov’t. FEBS Lett. 2016;590(12):1729–1738. doi:10.1002/1873-3468.12220
20. Xing Y, Wen X, Ding X, et al. CANT1 lncRNA triggers efficient therapeutic efficacy by correcting aberrant lncing cascade in malignant uveal melanoma. Research Support, Non-U.S. Gov’t. Mol Ther. 2017;25(5):1209–1221. doi:10.1016/j.ymthe.2017.02.016
21. Coupland SE, Lake SL, Zeschnigk M, Damato BE. Molecular pathology of uveal melanoma. Review. Eye (Lond). 2013;27(2):230–242. doi:10.1038/eye.2012.255
22. Li D, Lu J, Li H, Qi S, Yu L. Identification of a long noncoding RNA signature to predict outcomes of glioblastoma. Mol Med Rep. 2019;19(6):5406–5416. doi:10.3892/mmr.2019.10184
23. Nam S, Xie J, Perkins A, et al. Novel synthetic derivatives of the natural product berbamine inhibit Jak2/Stat3 signaling and induce apoptosis of human melanoma cells. Research Support, N.I.H., Extramural. Mol Oncol. 2012;6(5):484–493. doi:10.1016/j.molonc.2012.05.002
24. Zhong Z, Wen Z, Darnell JE
25. Sun X, Cao N, Mu L, Cao W. Stress induced phosphoprotein 1 promotes tumor growth and metastasis of melanoma via modulating JAK2/STAT3 pathway. Biomed Pharm. 2019;116:108962. doi:10.1016/j.biopha.2019.108962
26. Bousoik E, Montazeri Aliabadi H. “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway. Review. Front Oncol. 2018;8:287. doi:10.3389/fonc.2018.00287
27. Fu XQ, Chou JY, Li T, et al. The JAK2/STAT3 pathway is involved in the anti-melanoma effects of atractylenolide I. Letter Research Support, Non-U.S. Gov’t. Exp Dermatol. 2018;27(2):201–204. doi:10.1111/exd.13454
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.