Back to Journals » OncoTargets and Therapy » Volume 13
Knockdown of LncRNA DLEU2 Inhibits Cervical Cancer Progression via Targeting miR-128-3p
Authors Wang B, Hang J , Li W, Yuan W
Received 15 July 2020
Accepted for publication 11 September 2020
Published 9 October 2020 Volume 2020:13 Pages 10173—10184
DOI https://doi.org/10.2147/OTT.S272292
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Federico Perche
Bofei Wang,1 Jing Hang,2– 4 Weiling Li,5 Wanqiong Yuan6,7
1Department of Obstetrics and Gynecology, Weifang NO.2 People’s Hospital; 2Center for Reproductive Medicine, Department of Obstetrics and Gynecology, Beijing, People’s Republic of China; 3Peking University Third Hospital, Key Laboratory of Assisted Reproduction, Ministry of Education, Beijing, People’s Republic of China; 4Beijing Key Laboratory of Reproductive Endocrinology and Assisted Reproduction, Beijing, People’s Republic of China; 5Department of Obstetrics and Gynecology, Affiliated Yixing Hospital of Jiangsu University, Jiangsu, People’s Republic of China; 6Department of Orthopedics, Peking University Third Hospital, Beijing, People’s Republic of China; 7Beijing Key Laboratory of Spinal Disease, Beijing, People’s Republic of China
Correspondence: Weiling Li
Department of Obstetrics and Gynecology, Affiliated Yixing Hospital of Jiangsu University, No. 75. Tongzhenguan Road, Yixing, People’s Republic of China
Tel +86-13921327907
Email [email protected]
Wanqiong Yuan
Department of Orthopedics Peking University Third Hospital, 49 North Garden Road, Haidian District, Beijing, People’s Republic of China
Tel +86-15120092324
Fax +86 10-82266611
Email [email protected]
Objective: Cervical cancer is one of the most common female malignancies worldwide and represents a major global health challenge. The fast growth of tumor and high rates of metastasis still lead to a poor prognosis of cervical cancer patients. It is urgent to clarify the mechanism and identify predictive biomarkers for the treatment of cervical cancer. Long non-coding RNAs (LncRNAs) have been identified in cervical cancer and are related to malignant phenotypes of cervical cancer cells. However, the roles and mechanism of LncRNA deleted in lymphocytic leukemia (DLEU2) in the tumorigenesis and progression of cervical cancer remain unknown.
Materials and Methods: qPCR was performed to analyze the expression of DLEU2, Cyclin D1, CDK4, Bax, Bcl2 and mi-128-3p. Western blot was performed to detect the cell cycle hallmarks expression. CCK8 was used to examine cell proliferation. Cellular apoptosis was analyzed by Hoechst 33,258 staining and AV/PI staining with flow cytometry. Cell cycle was analyzed by flow cytometry. The xenograft model in nude mice was used to elucidate the function of DLEU2 in vivo. Bioinformatics analysis and luciferase reporter assay were proceeded to clarify whether miR-128-3p directly binds with lncRNA DLEU2. Pull‑down assay and RNA-binding protein immunoprecipitation assay were used for exploring the relationship between DLEU2 and miR-128-3p.
Results: We demonstrated that DLEU2 was upregulated in cervical cancer tumor tissues. Downregulation of DLEU2 inhibited cell proliferation, induced apoptosis and cell cycle arrest at G2/M phase of cervical cancer cells in vitro, and suppressed tumor growth in vivo. Further, LncRNA DLEU2 is one of the targets of miR-128-3p. miR-128-3p inhibitor abrogated the cell proliferation suppressed by knockdown of DLEU2, apoptosis induced by knockdown of DLEU2 and reversed the expression of cell cycle hallmarks regulated by knockdown of DLEU2.
Conclusion: Taken together, these results suggested knockdown of DLEU2 inhibited cervical cancer progression via targeting miR-128-3p.
Keywords: cervical cancer, LncRNAs, DLEU2, miR-128-3p, tumor growth
Introduction
Cervical cancer is one of the most common female malignancies worldwide and represents a major global health challenge with 570,000 cases and more than 300,000 deaths,1 hence becoming a disease threatening the health of females. It has been reported that cervical cancer patients have a poor prognosis because of the fast growth of tumor, high rates of lymphatic metastasis, distant metastases and low survival rate.2 Persistent infection of high-risk human papillomaviruses (HPVs) causes almost all cervical cancers.3,4 Although HPV vaccination is effective in preventing HPVs infection, it can not cure the identified infections. At present, the most commonly used treatments for cervical cancer are mainly based on operation, chemotherapy and radiotherapy. However, concomitant severe systemic toxicity and drug resistance of cervical cancer cells against the therapeutic agents cause poor therapeutic effects in cervical cancer.5 It is urgent to identify predictive biomarkers and therapeutic targets for treatment of cervical cancer and to clarify the mechanism that underlies the occurrence and progression of cervical cancer.
Long non-coding RNAs (LncRNAs) are broadly defined as transcripts longer than 200 bp nucleotides without protein-coding potential, including antisense, intronic, intergenic, pseudogene, and retrotransposon transcription.6 The aberrant expressions of LncRNAs are associated with many development processes of biological events, such as DNA methylation,7 histone modification,8,9 transcription and transcription regulation.10 LncRNAs may serve as the precursor of microRNAs (miRNAs),11,12 which construct an axis with LncRNAs to influence tumorigenesis and development.13–15 Recently, LncRNAs have received considerable attention because of their potential to regulate tumor progression.16 Abnormally expressed LncRNAs have been identified in cervical cancer and are related to malignant phenotypes of cervical cancer cells.17,18
The LncRNA deleted in lymphocytic leukemia (DLEU2) is transcribed from chromosome 13q14.319 with 15 exons.20 It has been reported that LncRNA DLEU2 serves as a tumor suppressor in several cancers, such as chronic lymphocytic leukemia21 and non-small cell lung cancer,22 which suggested that lncRNA DLEU2 may serve as a molecular biomarker for cancer diagnosis and treatment. However, the roles of DLEU2 in cervical cancer remain unknown. miRNA is a class of about 22 nucleotides conserved non-coding small RNA23,24 regulating developmental timing,25 cell proliferation, and apoptosis as well as tumor development.26–28 Aberrant miRNA expression is well recognized as a cancer hallmark. It has been reported that some miRNAs involved in the progression and development of cervical cancer, including miR-128,29 miR-30a,30 miR-224-3p31 and miR-186.32,33 Considering the interaction of lncRNA and miRNA plays an important role in cancer occurrence and progression,34 it is necessary to clarify the mechanism of lncRNA DLEU2 in cervical cancer by regulating miRNA in cervical cancer.
In this study, we found that DLEU2 was upregulated in tumor tissues of cervical cancer patients. Knockdown of DLEU2 inhibited the proliferation, induced apoptosis and cell arrest of cervical cancer cells in vitro and suppressed tumor growth in vivo. Further, LncRNA DLEU2 is one of the targets of miR-128-3p. MiR-128-3p inhibitor abrogated the tumor-suppressing properties induced by knockdown of DLEU2, including cell proliferation, apoptosis and the expression of cell cycle regulators. Taken together, we demonstrated that knockdown of DLEU2 inhibited cervical cancer via progression via targeting miR-128-3p.
Materials and Methods
Patient and Tissue Samples
The study cohort consisted of 50 tumor tissues and their paired distant normal tissues (obtained at least 5 cm from the surgical margins) collected from the cervical cancer patients that were diagnosed at Weifang NO.2 People’s Hospital (Shandong, China) from July 2019 to July 2020. Approval from the Research Ethics Committee of Weifang NO.2 People’s Hospital. None of the patients received neoadjuvant chemotherapy or radiation therapy. Fresh human tissues were frozen in liquid nitrogen for RNA extraction.
Cell Lines and Cell Cultures
The cervical cancer cell lines including HeLa, SiHa, C-33A and CaSKi and the human normal epithelial cell line HaCaT were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). These cells were cultured in high-glucose DMEM (Gibco, USA) with 10% fetal bovine serum (Biological Industries, Israel) at 37 °C with a humidified 5% CO2 environment.
Reverse Transcription (RT) and Quantitative Polymerase Chain Reaction (qPCR)
Total RNA from the tissues and cells was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was reverse-transcribed using a First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions for cDNA synthesis. All reactions were performed in three independent trials. The results were calculated using the 2-ΔΔCt method. U6 was the internal control for miR-128-3p and GAPDH was the internal control for other molecules. RT-qPCR was performed using the following primers as shown in Table 1.
 |
Table 1 Primer Sequences for qPCR |
Western Blot
Cells were washed with phosphate-buffered saline before total protein was extracted with radioimmunoprecipitation assay lysate (Beyotime, Shanghai, China) supplemented with protease inhibitor cocktail (Roche, Basel, Switzerland). Forty micrograms of entire protein lysates was split by SDS-PAGE. The protein was transferred into a PVDF membrane (Millipore, Bedford, MA, USA). Following overnight incubation with primary antibody Bcl-2 (Cat, 3498), CyclinD1 (Cat, 55,506), CDK4 (Cat, 12,790), Bax (Cat, 5023) and GAPDH (Cat, 5174) at 4°C. Then, the membranes were incubated with horseradish peroxidase-labeled secondary antibody at room temperature for 1 h. Signals were detected by LAS500 (GE, New York City, NY). All primary antibodies were purchased from CST (MA, USA) and secondary antibodies were purchased from Abcam (Cambridge, MA, USA).
siRNA Transfection
To knock down the endogenous DLEU2, siRNA pool (a mixture of following two target sequences with a proportion of 1 to 1: sequence 1#: 5ʹ-TTGCTGAAACTGCACAAAAAATC-3ʹ and sequence 2#: 5ʹ-TGCTGAAACTGCACAAAAAATCG-3ʹ) was transfected into cells. siRNA-negative control (si-NC) was used as a negative control: sequence 5′-UUCUCCGAACGUGUCACGUTT-3′ (Gene Pharma Inc., Shanghai, China). Cells were seeded in the six-well plate and transfected with the siRNA pool at a final concentration of 50 nM by Lipofectamine 3000 transfection reagent (Life Technologies, Carlsbad, CA) for 48 hours for subsequent experiments.
CCK8 Assay
Cells were seeded in the 96-well plate with 1000 cells per well. Ten microliters of CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan) was added to each well at the appointed time points. After one and a half hours of incubation, the absorbance was measured at 450 nm on a microplate reader (Promega Corporation, Fitchburg, WI, USA).
Hoechst 33,258 Staining
Apoptotic cells were evaluated by morphologic observation using Hoechst 33,258 (Solarbio, Beijing, China) staining. Specifically, cells at logarithmic growth were seeded in 6-well plates by density of 1×105 per well. After transfected with siRNA for 2 days, cells were fixed with 4% paraformaldehyde for 20 min at room temperature, and subsequently stained with Hoechst 33,258 at 37°C for 30 min. Cells were observed with a fluorescence microscope (Nikon, Tokyo, Japan).
Apoptosis Analysis
Cells were collected and resuspended in binding buffer and stained with FITC-Annexin V and PI (eBioscience, USA) for 15 min in darkness at room temperature. Samples were examined using a BD FACS Calibur flow cytometer (BD Biosciences, USA).
Cell Cycle Analysis
Cells were collected and incubated in 70% cold ethyl alcohol for at least 30 min at 4 °C. After that, RNAse (100 μg/mL) and propidium iodide (0.1 mg/mL) were added. The distribution of cell cycle phases was determined using Cell Quest software (BD Biosciences, USA).
Luciferase Reporter Assays
To identify the relationship between lncRNA DLEU2, DLEU2 containing the predicted binding sites with miR-128-3p or with mutated binding sites were cloned into a pMiR-REPORT (Promega, Madison, WI, USA) between the SgfI and NotI sites. The mutation of lncRNA DLEU2 was generated using QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, San Diego, CA, USA). Then, cervical cancer cells were co-transfected with miR-NC or miR-128-3p mimic and pMiR-DLEU2 wild type (DLEU2 WT) or pMiR-DLEU2 mutant type (DLEU2 MUT) using Lipofectamine 3000 (Invitrogen, USA). After 48 h of transfection, luciferase activity of lncRNA DLEU2 was analyzed using the Luciferase Reporter Assay System (Promega, USA) according to the manufacturer’s protocol.
Pull‑down Assay
DLEU2 and negative control (NC) were biotinylated to be bio-DLEU2, and bio-NC by GenePharma Company (Shanghai, China). Next, bio-DLEU2 or bio-NC was incubated with Dynabeads M-280 Streptavidin (Invitrogen, USA) for 1 hour at 4 °C. Then, Hela and SiHa cells were dissolved in the soft lysis buffer plus 80 U/mL RNasin (Promega Madison, WI, USA) and the cell lysates were incubated with the RNA-bound beads. Finally, the beads were washed with buffer and miR-128-3p were quantified and analyzed by qPCR.
RNA-Binding Protein Immunoprecipitation (RIP)
An RNA-Binding Protein Immunoprecipitation Kit (Merck, Millipore) was purchased to perform a RIP assay to determine the binding between the DLEU2 and miR-128-3p. The procedure complied with the guidance of the manufacturer. Antibodies for RIP assays against Ago2 and IgG were purchased from Abcam (ab5072, rabbit polyclonal antibody, Cambridge, MA, USA). Cell lysate was incubated with anti-Ago2 antibody or IgG antibody at 4 °C for 6 h. Finally, the extracted RNA was subjected to miR-128-3p enrichment using RT-qPCR.
In vivo Xenograft Experiments
6–8 weeks-BALB/c nude female mice (Charles River, China) were utilized to perform the xenograft experiments. All animal protocols were approved by the Institutional Animal Care and Use Committee at the Weifang NO.2 People’s Hospital. To establish the xenograft mice model, 1×107 HeLa and SiHa cells transfected with the indicated siRNA pool using the in vivo transfection reagent and mice were subcutaneously inoculated into the right flank.
Mice were monitored daily and the tumor volume was measured each week by calipers. Tumor volume was calculated using the following formula: tumor volume (mm3)=(height)×(width)2/2. After 5 weeks, mice were sacrificed and tumors were dissected and weighed. Tumor tissues were collected and snap-frozen in liquid nitrogen and stored at −80 °C for subsequent analyses.
Statistical Analyses
All the bars or symbols in the graph represented the means±SD from at least three independent experiments. Statistical significance (P <0.05) was calculated by Student’s t-test. The X-test (Fisher’s exact test) was carried out to analyze the correlations between DLEU2 expression.
Results
DLEU2 is Upregulated in Tumor Tissues and Knockdown of DLEU2 Inhibits Cell Proliferation of Cervical Cancer
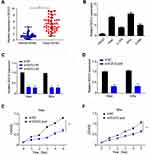
To fully understand the expression pattern of DLEU2 in cervical cancer, we analyzed the expression of DLEU2 in 50 paired cervical cancer tissues and their corresponding normal tissues. We observed that DLEU2 expression was upregulated in cervical cancer tissues compared with the paired normal tissues (Figure 1A), which suggested that DLEU2 might play crucial roles in the tumorigenesis and progress of cervical cancer.
To study the physiologic functions of DLEU2, we analyzed and compared its expression in HaCaT cells, an immortalized cervical epithelial cell line and cervical cancer cell lines, HeLa, SiHa, C-33A and CaSKi at the mRNA level by qPCR (Figure 1B) and chose HeLa and SiHa cells, which with higher DLEU2 expression levels to study the endogenous role of DLEU2. To avoid off-target effects, two siRNAs (si-DLEU2-1# and si-DLEU2-2#) were used to silence DLEU2 and the knockdown efficiency was typically more than 75% both in HeLa and SiHa cells (Figure 1C). In this paper, we used the siRNA pool (a mixture of the above two siRNAs with a proportion of 1 to 1) to study the endogenous roles of DLEU2 (Figure 1D). CCK8 assay was performed to analyze the effect of DLEU2 on cell proliferation. The results demonstrated that knockdown of DLEU2 significantly inhibited the proliferation of HeLa and SiHa cells compared with the si-NC cells (Figure 1E and F).
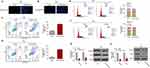
Knockdown of DLEU2 Induces Apoptosis and Cell Arrest of Cervical Cancer Cells
Next, the roles of DLEU2 in apoptosis and cell cycle were analyzed. Firstly, HeLa (Figure 2A) and SiHa (Figure 2B) were stained by Hoechst 33,258 after silencing DLEU2, and we observed that apoptosis occurred in DLEU2 knocked down cells. Then, Annexin V/PI staining further confirmed that knockdown of DLEU2 significantly induced apoptosis of HeLa and SiHa cells 48 h after transfection (Figure 2C and D). In addition, knockdown of DLEU2 induces cell cycle arrest at the G2/M phase compared with the control group in Hela and SiHa cells (Figure 2E and F). We further examined several key cell cycle regulators expression and found that Cyclin D1 and CDK expression were downregulated in the DLEU2-silenced HeLa (Figure 2G) and SiHa cells (Figure 2H) both at the mRNA level and protein level.
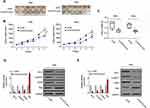
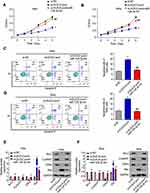
Knockdown of DLEU2 Suppresses Tumor Growth in vivo
In vitro experiments have shown that knockdown of DLEU2 suppressed cell proliferation, induced apoptosis and cell arrest at G2/M phase. In order to explore the effects of DLEU2 on tumor growth in vivo, a xenograft mice model was established by subcutaneous inoculation with HeLa-si-NC or HeLa-si-DLEU2 pool (Figure 3A, left) cells or SiHa-si-NC or SiHa-si-DLEU2-pool (Figure 3A, right) cells into the right flank of nude mice. The tumor growth was significantly inhibited in the DLEU2 silenced group mice compared with the si-NC group mice 5 weeks after inoculation. Besides, the tumor volume and weight of the si-DLEU2 pool mice were significantly reduced than those of the si-NC group mice (Figure 3B and C). Meanwhile, we detected the expression of Bcl-2, Cyclin D1, CDK4 and Bax with the tumors from xenograft mice by qPCR and Western blot and found that the expression of Bcl-2, Cyclin D1, and CDK4 was decreased in the tumors of DLEU2 silenced group mice and the expression of Bax was upregulated in si-DLEU2 pool-mice compared with the si-NC mice (Figure 3D and E).
LncRNA DLEU2 is One of the Targets of miR-128-3p
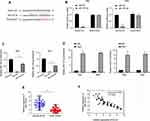
Increasing evidence have manifested that lncRNAs could function as “miRNA sponge” to bind miRNAs.11,12 We analyzed the relationship between DLEU2 and several miRNAs that may involve with the help of the bioinformatics online database (Starbase), including miR-128-3p, miR-30c-5p, miR-224-3p and miR-186. We found only that miR-128-3p has a potential binding site targeting lncRNA DLEU2 (Figure 4A). The luciferase reporter gene assays were performed to quantify these results as shown in Figure 4B in Hela and SiHa cells. Further, the RNA pull-down assay showed that there was a relationship between DLEU2 and miR-128-3p (Figure 4C). RIP assay indicated that higher enrichment of DLEU2 and miR-128-3p in anti-Ago2 relative to anti-IgG, confirming the interaction between DLEU2 and miR-128-3p in cervical cancer cells (Figure 4D). Moreover, we observed that the expression of miR-128-3p was downregulated in 50 cervical cancer tissues compared with the paired normal tissues (Figure 4E) and the expression of miR-128-3p was negatively correlated to DLEU2 expression (Figure 4F). Together, these changes suggested that LncRNA DLEU2 is one of the targets of miR-128-3p.
Knockdown of LncRNA DLEU2 Inhibits Cervical Cancer Progression in a miR-128-3p-Dependent Manner
To elucidate whether the inhibitory role of LncRNA DLEU2 in cervical cancer progression was dependent on miR-128-3p. The miR-128-3p inhibitor was utilized in si-DLEU2-pool cells and this treatment in si-DLEU2 pool cells reversed DLEU2-knockdown inhibited cell proliferation in Hela (Figure 5A) and SiHa cells (Figure 5B). Besides, miR-128-3p inhibitor in si-DLEU2 pool cells could also abolish the apoptosis induced by DLEU2 knockdown in Hela (Figure 5C) and SiHa (Figure 5D) cells. The expression of cell cycle regulators Bcl-2, Cyclin D1, and CDK4 was upregulated, and Bax was downregulated with treatment of miR-128-3p inhibitor in si-DLEU2 pool cells compared with the si-DLEU2 pool cells at mRNA level by qPCR and protein level by Western blot in Hela (Figure 5E) and SiHa (Figure 5F) cells. Taken together, the tumor-suppressive effects after the knockdown of DLEU2 were dependent on targeting miR-128-3p.
Discussion
Cervical cancer is a common malignancy that poses a prominent public health issue for women world while. At present, surgery, chemotherapy and radiotherapy therapy are the main strategies for the treatment of cervical cancer.35 However, it is regrettable that limitations still exist and a variety of side effects restrict the extensive application. Nowadays, although the proposal of molecular targeted therapies provides new treatment for cervical cancers, it is difficult to screen up a universal target for all types of cervical cancer.36 In this study, we aim to determine potential targets for cervical cancer treatment and found that DLEU2 was upregulated in cervical cancer patients. Knockdown of DLEU2 inhibited the proliferation, induced apoptosis and cell arrest at G2/M phase of cervical cancer cells in vitro and suppressed tumor growth in vivo. Mechanism study showed that there is an interaction between DLEU2 and miR-128-3p, and knockdown of DLEU2 suppressed cervical cancer progression via targeting miR-128-3p.
LncRNAs have been shown to regulate tumorigenesis, metastasis and involved in the therapy in several types of cancer,37 such as gastric cancer,38 lung cancer39 and breast cancer.40,41 Currently, a number of abnormally expressed LncRNAs have been identified in cervical cancer and participated in progress of cervical cancer.42,43 DLEU2 encodes a long noncoding RNA that is polyadenylated and spliced.44 The function of LncRNA DLEU2 in suppressing tumorigenesis and progression was reported recently, including esophageal cancer,45 hepatocellular carcinoma,46 pancreatic cancer,47 laryngeal squamous cell carcinoma48 and osteosarcoma.49 On the contrary, some reports suggested that a pro-oncogenic role of DLEU2 in cancers, such as clear cell renal cell carcinoma.50 These studies suggested that DLEU2 might show different functions in different cancer types due to the tissue specificity. LncRNAs might function as molecular sponges in modulating miRNAs.11,12 With the help of the bioinformatics online database, we found that miR-128-3p has a potential binding site targeting lncRNA DLEU2. In this study, we first showed the suppressive role of LncRNA DLEU2 via regulating miR-128-3p in cervical cancer. This observation provides more insights into the role of LncRNA DLEU2 in cervical cancer progression.
MicroRNAs are a type of small non-coding RNAs that can regulate various cellular processes via binding different target genes.23,24 They have been found to be dysregulated in cancer tissues compared to their matched normal tissues. miR-128-3p, known as miR-128, is the same major mature microRNA of miR-128-1 and miR-128-2, which are located on chromosomes 2q and 3q, respectively.51 miR-128-3p has been shown to be a key regulator in tumorigenesis and cancer development.52,53 Recently, Yao’s lab revealed that miR-128 was down-regulated in cervical cancer and negatively correlated with TNM stage, metastasis, tumor size, and poor prognosis of cervical cancer,28 which suggested that miR-128 played a role in cervical cancer. In our study, we proposed the mechanisms underlying the suppressive effects of LncRNA DLEU2 on cervical cancer growth via targeting miR-128-3p. It is necessary and interesting to explore the role of LncRNA DLEU2 in metastasis of cervical cancer in the future.
Conclusions
In this paper, we show that DLEU2 expression is upregulated in cervical cancer tissues compared with the paired normal tissues. Downregulation of DLEU2 inhibits the proliferation, induces apoptosis and cell arrest in G2/M phase in HeLa and SiHa cells in vitro and suppresses tumor growth in vivo. Further, knockdown of DLEU2 downregulates Bcl-2, Cyclin D1, and CDK4 expression, and upregulates Bax expression. Finally, we conclude LncRNA DLEU2 is one of the targets of miR-128-3p and knockdown of LncRNA DLEU2 inhibits cervical cancer progression via targeting miR-128-3p.
Abbreviations
LncRNAs, long non-coding RNAs; DLEU2, deleted in lymphocytic leukemia; HPVs, high-risk human papillomaviruses; RT, reverse transcription; qPCR, quantitative polymerase chain reaction; RIP, RNA-binding protein immunoprecipitation.
Data Sharing Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
All research studies on humans have been performed in accordance with the principles stated in the Declaration of Helsinki. This research was approved by the Ethical committee of Weifang NO.2 People’s Hospital (Approval Number: KT-E-2019-1-08). Informed consent was obtained from each patient. Experiments involving mice were performed in accordance with NC3Rs primate’s guidelines.
Consent for Publication
The authors agree with publication of this paper.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Key R&D Program of China (Grant Nos. 2018YFC1003702, 2018YFC1004403) and the Key Clinical Projects of Peking University Third Hospital (Grant No. BYSYZD2019041).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi:10.1016/S0140-6736(18)32470-X
3. Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382(9895):889–899.
4. Crow JM. HPV: the global burden. Nature. 2012;488(7413):S2–3. doi:10.1038/488S2a
5. Marth C, Landoni F, Mahner S, McCormack M, Gonzalez-Martin A, Colombo N. Cervical cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Suppl 4):iv262.
6. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
7. Ji J, Zhao L, Zhao X, et al. Genomewide DNA methylation regulation analysis of long noncoding RNAs in glioblastoma. Int J Mol Med. 2020;46(1):224–238.
8. Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics-Us. 2015;10(2):103–121. doi:10.1080/15592294.2014.1003746
9. Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37(1):3–9.
10. Zhang X, Wang W, Zhu W, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20(22):5573. doi:10.3390/ijms20225573
11. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14.
12. Tan JY, Sirey T, Honti F, et al. Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. 2015;25(5):655–666. doi:10.1101/gr.181974.114
13. Tong Y, Ru B, Zhang J, Kelso J. miRNA cancer MAP: an integrative web server inferring miRNA regulation network for cancer. Bioinformatics. 2018;34(18):3211–3213. doi:10.1093/bioinformatics/bty320
14. Li L, Peng M, Xue W, et al. Integrated analysis of dysregulated long non-coding RNAs/microRNAs/mRNAs in metastasis of lung adenocarcinoma. J Transl Med. 2018;16(1):372. doi:10.1186/s12967-018-1732-z
15. Tran D, Kessler C, Niehus SE, Mahnkopf M, Koch A, Tamura T. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene. 2018;37(1):75–85.
16. Dong P, Xiong Y, Yue J, et al. Long non-coding RNA NEAT1: a novel target for diagnosis and therapy in human tumors. Front Genet. 2018;9:471. doi:10.3389/fgene.2018.00471
17. Shang C, Wang W, Liao Y, et al. LNMICC promotes nodal metastasis of cervical cancer by reprogramming fatty acid metabolism. Cancer Res. 2018;78(4):877–890. doi:10.1158/0008-5472.CAN-17-2356
18. Yu X, Yang Y, Li Y, et al. Baicalein inhibits cervical cancer progression via downregulating long noncoding RNA BDLNR and its downstream PI3K/Akt pathway. Int J Biochem Cell Biol. 2018;94:107–118. doi:10.1016/j.biocel.2017.11.009
19. Dowd AA, Homeida S, Elkarem HA. Detection of chromosome 13 (13q14) deletion among Sudanese patients with multiple myeloma using a molecular genetics fluorescent in situ hybridization technique (FISH). Malays J Pathol. 2015;37(2):95–100.
20. Corcoran MM, Hammarsund M, Zhu C, et al. DLEU2 encodes an antisense RNA for the putative bicistronic RFP2/LEU5 gene in humans and mouse. Genes Chromosomes Cancer. 2004;40(4):285–297. doi:10.1002/gcc.20046
21. Klein U, Lia M, Crespo M, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi:10.1016/j.ccr.2009.11.019
22. Chen CQ, Chen CS, Chen JJ, et al. Histone deacetylases inhibitor trichostatin A increases the expression of Dleu2/miR-15a/16-1 via HDAC3 in non-small cell lung cancer. Mol Cell Biochem. 2013;383(1–2):137–148. doi:10.1007/s11010-013-1762-z
23. Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910.
24. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi:10.1038/nrg1379
25. Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21(4):511–517.
26. Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–110. doi:10.1038/nature13905
27. Tominaga N, Kosaka N, Ono M, et al. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun. 2015;6:6716. doi:10.1038/ncomms7716
28. Phuah NH, Azmi MN, Awang K, Nagoor NH. Suppression of microRNA-629 enhances sensitivity of cervical cancer cells to 1’S-1ʹ-acetoxychavicol acetate via regulating RSU1. Onco Targets Ther. 2017;10:1695–1705. doi:10.2147/OTT.S117492
29. Wu W, Guo L, Liang Z, Liu Y, Yao Z. Lnc-SNHG16/miR-128 axis modulates malignant phenotype through WNT/beta-catenin pathway in cervical cancer cells. J Cancer. 2020;11(8):2201–2212. doi:10.7150/jca.40319
30. Zhao J, Li B, Shu C, Ma Y, Gong Y. Downregulation of miR-30a is associated with proliferation and invasion via targeting MEF2D in cervical cancer. Oncol Lett. 2017;14(6):7437–7442.
31. Fang W, Shu S, Yongmei L, Endong Z, Lirong Y, Bei S. miR-224-3p inhibits autophagy in cervical cancer cells by targeting FIP200. Sci Rep. 2016;6:33229. doi:10.1038/srep33229
32. Liu C, Wang J, Hu Y, Xie H, Liu M, Tang H. Upregulation of kazrin F by miR-186 suppresses apoptosis but promotes epithelial-mesenchymal transition to contribute to malignancy in human cervical cancer cells. Chin J Cancer Res. 2017;29(1):45–56. doi:10.21147/j.issn.1000-9604.2017.01.06
33. Liu Y, Guo R, Qiao Y, Han L, Liu M. LncRNA NNT-AS1 contributes to the cisplatin resistance of cervical cancer through NNT-AS1/miR-186/HMGB1 axis. Cancer Cell Int. 2020;20:190. doi:10.1186/s12935-020-01278-9
34. Janakiraman H, House RP, Gangaraju VK, Diehl JA, Howe PH, Palanisamy V. The long (lncRNA) and short (miRNA) of it: TGFbeta-mediated control of RNA-binding proteins and noncoding RNAs. Mol Cancer Res. 2018;16(4):567–579. doi:10.1158/1541-7786.MCR-17-0547
35. Small WJ, Bacon MA, Bajaj A, et al. Cervical cancer: a global health crisis. Cancer-Am Cancer Soc. 2017;123(13):2404–2412.
36. Zhu H, Luo H, Zhang W, Shen Z, Hu X, Zhu X. Molecular mechanisms of cisplatin resistance in cervical cancer. Drug Des Devel Ther. 2016;10:1885–1895. doi:10.2147/DDDT.S106412
37. Farooqi AA, Attar R, Qureshi MZ, et al. Interplay of long non-coding RNAs and TGF/SMAD signaling in different cancers. Cell Mol Biol (Noisy-Le-Grand). 2018;64(15):1–6.
38. Duan F, Jiang J, Song C, et al. Functional long non-coding RNAs associated with gastric cancer susceptibility and evaluation of the epidemiological efficacy in a central Chinese population. Gene. 2018;646:227–233. doi:10.1016/j.gene.2017.12.063
39. Jiang R, Tang J, Chen Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129. doi:10.1038/ncomms15129
40. Guan YX, Zhang MZ, Chen XZ, Zhang Q, Liu SZ, Zhang YL. Lnc RNA SNHG20 participated in proliferation, invasion, and migration of breast cancer cells via miR-495. J Cell Biochem. 2018;119(10):7971–7981. doi:10.1002/jcb.26588
41. Brown JM, Wasson MD, Marcato P. The missing Lnc: the potential of targeting triple-negative breast cancer and cancer stem cells by inhibiting long non-coding RNAs. Cells-Basel. 2020;9(3).
42. Su K, Zhao Q, Bian A, Wang C, Cai Y, Zhang Y. A novel positive feedback regulation between long noncoding RNA UICC and IL-6/STAT3 signaling promotes cervical cancer progression. Am J Cancer Res. 2018;8(7):1176–1189.
43. Wei H, Qiu YQ, Zeng QS, Wang SF, Yi CJ. LncRNA UCA1 regulates proliferation, migration and invasion of cervical cancer cells by targeting miR-145. Eur Rev Med Pharmacol Sci. 2020;24(7):3555–3564.
44. Migliazza A, Bosch F, Komatsu H, et al. Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood. 2001;97(7):2098–2104. doi:10.1182/blood.V97.7.2098
45. Lu T, Wang R, Cai H, Cui Y. Long non-coding RNA DLEU2 promotes the progression of esophageal cancer through miR-30e-5p/E2F7 axis. Biomed Pharmacother. 2020;123:109650.
46. Guo Y, Bai M, Lin L, et al. LncRNA DLEU2 aggravates the progression of hepatocellular carcinoma through binding to EZH2. Biomed Pharmacother. 2019;118:109272. doi:10.1016/j.biopha.2019.109272
47. Xu B, Gong X, Zi L, et al. Silencing of DLEU2 suppresses pancreatic cancer cell proliferation and invasion by upregulating microRNA-455. Cancer Sci. 2019;110(5):1676–1685. doi:10.1111/cas.13987
48. Li X, Xu F, Meng Q, et al. Long noncoding RNA DLEU2 predicts a poor prognosis and enhances malignant properties in laryngeal squamous cell carcinoma through the miR-30c-5p/PIK3CD/Akt axis. Cell Death Dis. 2020;11(6):472. doi:10.1038/s41419-020-2581-2
49. Liu W, Liu PC, Ma K, Wang YY, Chi QB, Yan M. LncRNA DLEU2 promotes tumour growth by sponging miR-337-3p in human osteosarcoma. Cell Biochem Funct. 2020.
50. Chen Z, Zhang J, Zhang Z, et al. The putative tumor suppressor microRNA-30a-5p modulates clear cell renal cell carcinoma aggressiveness through repression of ZEB2. Cell Death Dis. 2017;8(6):e2859. doi:10.1038/cddis.2017.252
51. Hu J, Cheng Y, Li Y, et al. microRNA-128 plays a critical role in human non-small cell lung cancer tumourigenesis, angiogenesis and lymphangiogenesis by directly targeting vascular endothelial growth factor-C. Eur J Cancer. 2014;50(13):2336–2350. doi:10.1016/j.ejca.2014.06.005
52. Liu T, Zhang X, Du L, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18(1):43.
53. Cai J, Fang L, Huang Y, et al. Simultaneous overactivation of Wnt/beta-catenin and TGFbeta signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat Commun. 2017;8:15870.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.