Back to Journals » OncoTargets and Therapy » Volume 13
Knockdown of circ_0000512 Inhibits Cell Proliferation and Promotes Apoptosis in Colorectal Cancer by Regulating miR-296-5p/RUNX1 Axis
Authors Wang L, Wu H, Chu F, Zhang L, Xiao X
Received 19 February 2020
Accepted for publication 3 July 2020
Published 28 July 2020 Volume 2020:13 Pages 7357—7368
DOI https://doi.org/10.2147/OTT.S250495
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Lihong Wang, Huili Wu, Feifei Chu, Li Zhang, Xingguo Xiao
Department of Gastroenterology, Zhengzhou Central Hospital Affiliated to Zhengzhou University, Zhengzhou 450000, Henan, People’s Republic of China
Correspondence: Huili Wu Email [email protected]
Background: Colorectal cancer (CRC) is one of the leading causes of cancer-related death worldwide. Increasing evidence showed that circular RNAs (circRNAs) played critical roles in the progression of CRC. However, the effects and underlying mechanisms of circ_0000512 in CRC progression remain unclear.
Methods: The expression levels of circ_0000512, microRNA-296-5p (miR-296-5p) and runt-related transcription factor 1 (RUNX1) were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). Cell viability, colony formation, cell cycle distribution and cell apoptosis were detected by Cell Counting Kit-8 (CCK-8) assay, colony formation assay and flow cytometry analysis, respectively. Western blot assay was utilized to measure the protein expression of Cyclin D1, Cleaved Caspase-3 and RUNX1. The interaction between miR-296-5p and circ_0000512 or RUNX1 was predicted by starBase and verified by dual-luciferase reporter assay, RNA immunoprecipitation (RIP) assay and RNA pull-down assay. The mice xenograft model was established to explore the function of circ_0000512 in vivo.
Results: The expression of circ_0000512 was increased in CRC tissues and cells. Knockdown of circ_0000512 suppressed cell viability and colony formation and arrested the cells at the G0/G1 phase while it accelerated apoptosis in CRC cells. Mechanistically, circ_0000512 could increase RUNX1 expression by acting as a molecular sponge of miR-296-5p in CRC cells. Furthermore, miR-296-5p downregulation or RUNX1 overexpression reversed the anti-proliferation and pro-apoptosis effects caused by circ_0000512 knockdown in CRC cells. In addition, circ_0000512 interference inhibited tumor growth by upregulating miR-296-5p and downregulating RUNX1 in vivo.
Conclusion: Knockdown of circ_0000512 inhibited cell proliferation and induced apoptosis in CRC cell by regulating miR-296-5p/RUNX1 axis, which might provide a potential therapeutic target for CRC treatment.
Keywords: colorectal cancer, circ_0000512, -miR-296-5p, RUNX1, proliferation, apoptosis
Introduction
Colorectal cancer (CRC) is one of the most lethal and prevalent cancers worldwide, with an estimated 1.8 million cases and 881000 deaths in 2018.1 Despite great advancement in therapeutic approaches, such as surgery, chemotherapy and radiation therapy, the overall survival rate of CRC patients is still unsatisfactory.2 Hence, it is important to investigate its underlying mechanisms and discover new therapeutic targets for CRC treatment.
Circular RNAs (circRNAs), a special type of non-coding RNAs, have covalently closed-loop structures with neither 5ʹ cap nor 3ʹ polyadenylated tail (unlike linear RNAs).3 Owing to their closed-loop structures, circRNAs not easily affected by RNA exonuclease and more stable than linear RNAs.4 Accumulating evidence showed that circRNAs are widely expressed in various types of cells and participated in the tumorigenesis and development of many tumors, including CRC.5,6 For instance, hsa_circ_0136666 accelerated CRC cell growth and invasion via modulating miR-136/SH2B1 axis.7 Hsa_circ_000984 has been suggested to function as a positive regulator in the growth and metastasis of CRC cells through sponging miR-106b.8 As for hsa_circ_0000512 (circ_0000512; chr14:20811282–20811436; also known as hsa_circ_000166), it has been suggested to be overexpressed in CRC tissues.9 However, the underlying mechanism and biological function of circ_0000512 in CRC have not been reported.
Recent studies have shown that circRNAs usually act as competitive endogenous RNAs (ceRNAs; also known as microRNA sponges) to regulate target gene expression, thereby regulating cell behaviors, such as proliferation, metastasis and apoptosis.10,11 MicroRNAs (miRNAs) are a class of short non-coding RNAs that affect gene expression via promoting mRNA degradation or suppressing mRNA translation.12 MiR-296-5p was an anti-oncogene in several cancers, including CRC.13,14 However, the connection between circ_0000512 and miR-296-5p has not been elucidated. Runt-related transcription factor 1 (RUNX1), a transcription factor, functions as either a tumor promoter or tumor suppressor gene in epithelial tumors, and its oncogenic role has received more and more attention.15–18 RUNX1 has been demonstrated to act as an anti-oncogene in CRC.19 Bioinformatics analysis predicted the potential binding sites between miR-296-5p and circ_0000512 or RUNX1, implying that circ_0000512 might serve as a molecular sponge of miR-296-5p to modulate RUNX1 expression. Therefore, the relationships among circ_0000512, miR-296-5p and RUNX1 and their underlying mechanisms will be the focus of our study.
In this research, the expression of circ_0000512 was analyzed in CRC tissues and cells. Additionally, we explored the biological functions of circ_0000512 on proliferation, cell cycle process and apoptosis, and investigated the ceRNA regulatory network of circ_0000512/miR-296-5p/RUNX1. Our purpose was to provide a novel regulatory mechanism of circ_0000512 in the progression CRC and offer a theoretical basis for the treatment of CRC.
Materials and Methods
Tissue Collection
In this study, 55 pairs of CRC tissues and normal tissues were provided by the patients who had undergone surgery at Zhengzhou Central Hospital Affiliated to Zhengzhou University. After surgical resection, these samples were collected, promptly frozen in liquid nitrogen and then stored in a refrigerator at −80. All participants had signed the informed consents prior to collecting the tissues. All experiments conducted in the current study were granted by the Research Ethics Committee of Zhengzhou Central Hospital Affiliated to Zhengzhou University.
Cell Culture and Transfection
Normal colon FHC cells and four CRC cell lines (SW480, HCT116, SW620, LoVo) were purchased from COBIOER (Nanjing, China) and cultured in Dulbecco’s modified eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA), 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C under a humidified atmosphere containing 5% CO2.
Short hairpin RNA (shRNA) targeting circ_0000512 (sh-circ#1, sh-circ#2 and sh-circ#3) and corresponding negative control (sh-NC), RUNX1 overexpression vector (RUNX1) and empty vector (vector) were purchased from Genechem (Shanghai, China). MiR-296-5p mimic or inhibitor (miR-296-5p or anti-miR-296-5p) and their negative controls (miR-NC or anti-NC) were synthesized by GenePharma (Shanghai, China). HCT116 and SW620 cells were transfected with these oligonucleotides (50 nM) or vectors (2 μg) by Lipofectamine 3000 reagent (Invitrogen).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA from tissues and cells were isolated by Trizol reagent (Invitrogen). For detecting genes expression, the first strand of complementary DNA (cDNA) was synthesized by High-Capacity cDNA Reverse Transcription Kit and TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). Next, qRT-PCR analysis was conducted using the SYBR Premix Ex Taq (Takara, Dalian, China) on the ABI 7300 system (Thermo Fisher Scientific). The sequences of primers used in this research were listed as followed: circ_0000512 (Forward, 5ʹ-GGAACAGACTCACGGCCA-3ʹ; Reverse, 5ʹ-CATCTCCTGCCCAGTCTGAC-3ʹ); miR-296-5p (Forward, 5ʹ-TGCCTAATTCAGAGGGTTGG-3ʹ; Reverse, 5ʹ-CTCCACTCCTGGCACACAG-3ʹ); RUNX1 (Forward, 5ʹ-CGAAGACATCGGCAGAAACT-3ʹ; Reverse, 5ʹ-TAAAGGCAGTGGAGTGGTTCA-3ʹ); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Forward, 5ʹ-CGCTCTCTGCTCCTCCTGTTC-3ʹ; Reverse, 5ʹ- ATCCGTTGACTCCGACCTTCAC-3ʹ), U6 (Forward, 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ; Reverse, 5ʹ-ACGCTTCACGAATTTGCGTGTC-3ʹ). The circ_0000512, RUNX1 or miR-296-5p expression was evaluated with calculated using 2-ΔΔCt method with normalization to GAPDH or U6, respectively.
RNase R Treatment
To evaluate the stability of circ_0000512, total RNA (2 μg) was incubated with the RNase R (3 U/μg; Epicentre Technologies, Madison, WI, USA) for 30 min at 37°C. The cells were collected following treatment with RNase R, and qRT-PCR was performed for detecting the expression of circ_0000512 and GAPDH.
Cell Viability Assay
Cell Counting Kit-8 (CCK-8; Sangon Biotech, Shanghai, China) was used to detect cell viability. Briefly, HCT116 and SW620 cells were seeded in 96-well plates. CCK-8 (10 μL) solution was added to per well after transfection for 24 hrs, 48 hrs or 72 hrs. Afterward, the plates were placed in the incubator for 3 h and then the absorbance of per well was analyzed using a microplate reader (Bio-Rad, Hercules, CA, USA) at 450 nm.
Colony Formation Assay
Transfected HCT116 and SW620 cells were seeded in six-well plates and the medium was updated every 3 days. The medium was carefully removed after incubation for 14 days, and the cells were washed with phosphate-buffered saline (PBS). After that, cells were fixed using paraformaldehyde (4%). Colonies (one colony more than 50 cells) were counted using the microscope.
Cell Cycle Assay
HCT116 and SW620 cells were collected following transfection for 48 hrs, and fixed using ice‐cold ethanol (70%) for 24 h at −20°C. Next, the cells were collected and washed by PBS, and then stained with 25 μg/mL propidium iodide (PI) solution in PBS containing 0.2% Triton X‐100 and 50 μg/mL RNase for 20 min in the darkness. Lastly, flow cytometry (Guava Technologies, Hayward, CA, USA) was used to analyze the cell cycle distribution.
Cell Apoptosis Assay
Apoptotic cells were analyzed using the Annexin V-FITC/PI apoptosis detection kit (Sangon Biotech) following the standard protocol. Briefly, transfected HCT116 and SW620 cells were re-suspended in binding buffer (400 μL), and then stained with Annexin V-FITC (10 μL) and PI (5μL) for 15 min in the darkness. Lastly, the apoptotic cells were determined by flow cytometry.
Western Blot Assay
Total protein was extracted by RIPA lysis buffer (Sangon Biotech). After that, BCA protein assay kit (Tanon, Shanghai, China) was utilized to quantify the total protein. The protein (about 40 µg) was resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred onto the nitrocellulose membrane (Millipore, Billerica, MA, USA). The membranes were blocked with 5% nonfat dried milk (Yili, Beijing, China). Afterward, the membranes were probed by primary antibody against Cyclin D1 (1:1500, ab226977, Abcam, Cambridge, UK), Cleaved Caspase-3 (1:500, ab49822, Abcam), RUNX1 (1:500, ab23980, Abcam) or GAPDH (1:2500, ab9485, Abcam) at 4°C for 12 h, followed by interaction with HRP-conjugated anti-rabbit IgG (1:4000, ab205718, Abcam). Finally, enhanced chemiluminescence reagent (Tanon) was used to observe the protein bands, and ImageJ software was employed to quantify the protein expression.
Subcellular Fractionation Location
PARIS Kit (Life Technologies Corp., Grand Island, NY, USA) was utilized to isolate cytosolic and nuclear fractions. Briefly, HCT116 and SW620 were washed by PBS and placed on ice. Afterward, HCT116 and SW620 cells were re-suspended in fractionation buffer. After centrifugation at 500 × g at 4°C for 5 min, the cytoplasmic fraction was separated from the nuclear pellet. Then, the remaining nuclear pellet was again lysed using the cell disruption buffer as nuclear fraction. After that, the sample was divided for RNA isolation. Lastly, the abundance of U6, GAPDH and circ_0000512 was analyzed by qRT-PCR in the nuclear and cytoplasmic fractions. GAPDH and U6 were functioned as controls for the cytoplasmic and nuclear, respectively.
Dual-Luciferase Reporter Assay
The potential binding sites of miR-296-5p and circ_0000512 or RUNX1 were predicted by starBase v2.0. The circ_0000512 or RUNX1 fragments including wild-type (WT) or mutant (MUT) binding sites of miR-296-5p were synthesized and then cloned into pmirGlO luciferase reporter vector (Promega, Madison, WI, USA), namely WT plasmids (circ_0000512-WT, RUNX1 3ʹUTR-WT) or MUT plasmids (circ_0000512-MUT, RUNX1-MUT). Then, HCT116 and SW620 cells were co-transfected with constructed plasmid (WT or MUT) and miR-296-5p (or miR-NC). Finally, the luciferase activity was determined via a dual-luciferase reporter assay system (Promega) after transfection for 48 h.
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted using the Magna RIP Kit (Millipore). In short, HCT116 and SW620 cells were lysed using the RIP buffer. Subsequently, lysates were incubated in RIP buffer containing magnetic beads conjugated with anti-argonaute 2 (anti-ago2) or IgG antibody. After that, these samples were incubated by Proteinase K to digest the protein, followed by RNA purification. Afterward, the levels of circ_0000512 or miR-296-5p were examined using the qRT-PCR analysis as described above.
RNA Pull-Down Assay
RNA pull-down assay was conducted in HCT116 and SW620 cells using the RNA-Protein Pull-Down Kit (Thermo Fisher Scientific) to assess the interaction between miR-296-5p and circ_0000512. In short, miR-NC or miR-296-5p was labeled with biotin and transfected into HCT116 and SW620 cells. Cell lysates were incubated by streptavidin magnetic beads. Afterward, the complex was eluted using the biotin elution buffer, and circ_0000512 level was evaluated using the qRT-PCR assay.
In vivo Tumor Growth Assay
Sh-circ#2 or sh-NC was transfected into SW620 cells. Subsequently, stably transfected cells (3×106) were subcutaneously injected in BALB/c nude mice (female, 5-week-old, n=6/group, Huafukang, Beijing, China). Tumor volume was gauged using a caliper every 7 days and calculated using the formula: length × width2 ×0.5. These mice were sacrificed after injection for 35 days, and resected tumor masses were weighed and collected for detection of circ_0000512, miR-296-5p and RUNX1 levels. The animal experiments were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of “National Institutes of Health” and granted by the committee of Animal Research of Zhengzhou Central Hospital Affiliated to Zhengzhou University.
Statistical Analysis
In this study, all data from at least three independent experiments were displayed as mean ± standard deviation (SD). The significance of differences between groups was analyzed with Student’s t-test (for two groups) or one-way analysis of variance (ANOVA; for more than two groups). Kaplan–Meier method was applied to create a survival curve. All statistical analyses were conducted using the Graphpad Prism version 6.0 software (GraphPad Software, San Diego California, USA). P<0.05 exhibited a statistical significance.
Results
Circ_0000512 Was Upregulated in CRC Tissues and Cells
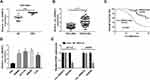
Firstly, we analyzed GSE126094 (10 pairs of CRC tissues and normal tissues) from the Gene Expression Omnibus (GEO) database to explore the relative expression of circ_0000512. The data showed that the expression of circ_0000512 was enhanced in CRC tissues compared to normal tissues (Figure 1A). To further confirm the expression of circ_0000512 in CRC tissues and cells, qRT-PCR was conducted. The results showed that the expression of circ_0000512 was increased in CRC tissues relative to normal tissues (Figure 1B). Moreover, we further divided 55 CRC patients into with high circ_0000512 (n=23) and low circ_0000512 (n=22) expression groups according to median value of circ_0000512 expression. As shown in Figure 1C, patients with high circ_0000512 expression group presented poor survival in contrast to the low expression group (P<0.001). In addition, we also found that the level of circ_0000512 was upregulated in CRC cells (SW480, HCT116, SW620, LoVo) compared to FHC cells (Figure 1D). Next, RNase R was utilized in the experiments to determine the stability of circ_0000512. Results revealed that circ_0000512 was resistant to RNase R compared with the linear GAPDH (Figure 1E), disclosing that circ_0000512 formed a loop structure. These results indicated that circ_0000512 might play critical roles in the progression of CRC.
Knockdown of circ_0000512 Inhibited Cell Proliferation and Induced Apoptosis in CRC Cells
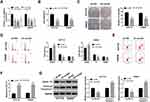
To investigate the potential biological function of circ_0000512 in CRC progression, we silenced circ_0000512 expression in HCT116 and SW620 cells by transfection circ_0000512 shRNA. As displayed in Figure 2A, the expression of circ_0000512 was obviously decreased in HCT116 and SW620 cells transfected with sh-circ#1, sh-circ#2 and sh-circ#3, and circ_0000512 expression was the lowest in the sh-circ#2 transfected cells. Thus, sh-circ#2 was used in subsequent experiments. CCK-8 assay indicated that interference of circ_0000512 inhibited the viability of HCT116 and SW620 cells (Figure 2B). Colony formation assay indicated that circ_0000512 knockdown reduced the number of colonies in HCT116 and SW620 cells (Figure 2C). Cell cycle progression was analyzed by flow cytometry. As depicted in Figure 2D, the percentage of G0/G1 phase cells was elevated by downregulating circ_0000512, while the percentage of cells in S phase was obviously reduced after the interference of circ_0000512, suggesting that the cell cycle was arrested at the G0/G1 phase. Moreover, we found that cell apoptosis was increased in HCT116 and SW620 cells transfected with sh-circ#2 compared to the sh-NC group (Figure 2E and F). Cyclin D1, a member of a family of three closely associated D-type cyclins, is required for cell cycle progression in G1 and is a growth-promoting protein.20,21 Western blot assay was performed to examine the protein levels of Cyclin D1 and Cleaved caspase-3 (a key executor in the apoptotic process). Results presented that silence of circ_0000512 led to a decrease in Cyclin D1 expression and an increase in Cleaved caspase-3 expression (Figure 2G). These data demonstrated that silencing circ_0000512 could inhibit the progression of CRC.
Circ_0000512 Served as a Sponge of miR-296-5p
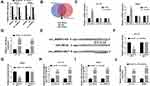
Next, the localization of circ_0000512 was analyzed in HCT116 and SW620 cells. The data revealed that most of the circ_0000512 was located in the cytoplasm (Figure 3A). Previous study reported that circRNAs could act as a molecular sponge to interact with miRNAs,22 so the potential target miRNAs of circ_0000512 were predicted by starBase and circinteractom. The results showed that miR-296-5p, miR-326 and miR-330-5p might be targets of circ_0000512 (Figure 3B). We found that miR-296-5p was significantly upregulated by downregulation of circ_0000512 while the expression of miR-326 and miR-330-5p was not evidently changed (Figure 3C), so we focused on the relationship between miR-296-5p and circ_0000512 in CRC cells. QRT-PCR analysis showed that the expression of miR-296-5p was markedly increased in HCT116 and SW620 cells transfected with miR-296-5p mimic (Figure 3D), suggesting that transfection of miR-296-5p was successful. Starbase showed the putative binding sites of miR-296-5p and circ_0000512 (Figure 3E). Next, the prediction was confirmed by dual-luciferase reporter, RIP and RNA pull-down analyses. Dual-luciferase reporter analysis showed that the luciferase activity of circ_0000512-WT was strikingly suppressed in cells transfected with miR-296-5p, while luciferase activity of circ_0000512-MUT was not changed (Figure 3F and G). In addition, RIP assay revealed that enrichment of miR-296-5p and circ_0000512 was conspicuously enhanced in anti-ago2 group compared with that in the anti-IgG group (Figure 3H and I). Moreover, Bio-miR-296-5p led to a higher circ_0000512 level than the treatment of Bio-NC in HCT116 and SW620 cells (Figure 3J). All these results demonstrated that circ_0000512 could function as a sponge of miR-296-5p in CRC.
RUNX1 Was a Downstream Target of miR-296-5p
To investigate the underlying mechanism of miR-296-5p, starBase was used and RUNX1 was identified as a potential downstream target of miR-296-5p (Figure 4A). The dual-luciferase reporter assay proved that transfection of miR-296-5p significantly reduced the luciferase activity of RUNX1 3ʹUTR-MUT, but not the luciferase activity of RUNX1 3ʹUTR-MUT in HCT116 and SW620 cells (Figure 4B). Subsequently, the regulatory effect of miR-296-5p on RUNX1 expression was explored. As illustrated in Figure 4C and D, the mRNA and protein expression of RUNX1 were prominently decreased after transfection of miR-296-5p in HCT116 and SW620 cells. QRT-PCR displayed that the abundance of miR-296-5p was evidently decreased in HCT116 and SW620 cells transfected with anti-miR-296-5p (Figure 4E), suggesting miR-296-5p was successfully knocked down. Next, we explored whether circ_0000512 functioned as a molecular sponge of miR-296-5p to regulate RUNX1 expression. We found that circ_0000512 silence decreased the mRNA and protein expression of RUNX1 while knockdown of miR-296-5p abated this effect (Figure 4F and G). To sum up, these findings elaborated that circ_0000512 regulated RUNX1 expression by sponging miR-296-5p in CRC cells.
Circ_0000512 Downregulation Suppressed the Progression of CRC Cells by Upregulating miR-296-5p and Downregulating RUNX1
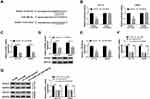
The transfection efficiency of RUNX1 was determined by Western blot assay. It was found that the protein abundance of RUNX1 was increased in HCT116 and SW620 cells transfected with RUNX1 (Figure 5A). We next analyzed whether the function of circ_0000512 in CRC cells was mediated by miR-296-5p and RUNX1. The inhibitory effects of circ_0000512 interference on cell viability and colony formation were abolished by the downregulation of miR-296-5p or upregulation of RUNX1 in HCT116 and SW620 cells (Figure 5B and C). Moreover, knockdown of miR-296-5p or overexpression of RUNX1 could reverse the circ_0000512 silence-mediated promotion of G0/G1 phase cells and reduction of S phase cells (Figure 5D and E). Likewise, the effects of circ_0000512 knockdown on inhibition of Cyclin D1 expression and enhancement of Cleaved caspase-3 expression were also abated by downregulating miR-296-5p or upregulating RUNX1 (Figure 5F). Taken together, these findings implied that circ_0000512 exerted its tumor oncogenic roles via regulating the miR-296-5p/RUNX1 axis in CRC cells.
Interference of circ_0000512 Restrained the Tumor Growth by Regulating miR-296-5p and RUNX1 Expression
To assess the effect of circ_0000512 on CRC in vivo, sh-NC or sh-circ#2-transfected SW620 cells were introduced into nude mice. As presented in Figure 6A and B, knockdown of circ_0000512 decreased tumor volume and weight in the xenograft model. Next, the expression of circ_0000512, miR-296-5p and RUNX1 was measured in tumor tissues. As expected, qRT-PCR demonstrated that knockdown of circ_0000512 decreased the expression of circ_0000512 and enhanced the abundance of miR-296-5p in excised tumor masses (Figure 6C and D). Western blot assay showed that interference of circ_0000512 resulted in a decrease of RUNX1 protein expression in tumor tissues (Figure 6E). The above results indicated that circ_0000512 downregulation inhibited tumor growth through upregulating miR-296-5p and downregulating RUNX1 in vivo.
Discussion
Many studies showed that dysregulation of circRNAs was closely associated with CRC tumorigenesis.23,24 Nevertheless, the effect of circ_0000512 on CRC progression has not been explored. Therefore, the purpose of our study was to explore the biological effect and molecular mechanism of circ_0000512 in CRC cells.
Growing evidence has certified that multiple circRNAs were abnormally expressed in diseases and recognized as significant prognostic biomarkers of many cancers.3,25 Besides, circRNAs could modulate the proliferation, metastasis and apoptosis of CRC cells through function as molecular sponges of miRNAs. For instance, Bian et al proved that hsa_circRNA_103809 could regulate CRC cell growth and metastasis via sponging miR-532-3P to modulate FOXO4 expression.26 Fang et al stated that circRNA_100290 level was enhanced in CRC tissues and cells, and its knockdown repressed CRC cell proliferation and metastasis but promoted apoptosis through sponging miR-516b.27 Geng et al9 and Chen et al28 pointed out that circ_0000512 abundance was enhanced in CRC tissues, implying that circ_0000512 might play a pivotal role in CRC progression. In line with these findings, we also found that circ_0000512 expression was elevated in CRC tissues and cells. The functional experiments presented that interference of circ_0000512 inhibited cell viability and colony formation and arrested cells at the G0/G1 phase while accelerated apoptosis in CRC cells. These findings suggested that circ_0000512 acted as an oncogene in CRC.
To explore whether circ_0000512 functioned as miRNA sponges to regulate target genes expression in CRC, bioinformatics software was used to predict the target miRNAs of circ_0000512. Results showed that miR-296-5p, miR-326 and miR-330-5p might be targets of circ_0000512. Subsequently, a series of experiments demonstrated that circ_0000512 could bind to miR-296-5p. It has been reported that miR-296-5p abundance was usually downregulated and acted as an anti-oncogene in many tumors, including prostate cancer,13 lung cancer29 and hepatocellular carcinoma.30 However, some studies suggested that miR-296-5p played an oncogenic role in gastric cancer31 and neuroblastoma.32 These findings disclosed that miR-296-5p played different roles in different cancers. Besides, He et al proved that miR-296 level was decreased in CRC tissues and cells, and suppressed CRC cell metastasis via targeting S100A4.33 Likewise, Zhang et al revealed that miR-296 upregulation repressed CRC cell growth and enhanced apoptosis through targeting ARRB1.14 As miRNAs exert their biological functions through modulating their downstream targets.34 Hence, the potential targets of miR-296-5p were predicted using the starBase software. The data presented that RUNX1 contained binding sites of miR-296-5p, which was then verified by dual-luciferase reporter assay in CRC cells. Moreover, we found that knockdown of circ_0000512 reduced RUNX1 expression, while this effect could be abated by downregulating miR-296-5p, suggesting that circ_0000512 acted as miR-296-5p sponge to positively regulate RUNX1 expression. RUNX1 has been reported to be overexpressed in CRC tissues and its overexpression contributed to metastasis via activating Wnt/β-catenin signaling pathway.19 Besides, Zhou et al found that interference of RUNX1 inhibited CRC proliferation and promoted apoptosis.35 Next, we explored the effect of miR-296-5p and RUNX1 on biological functions of circ_0000512 in CRC cells. We observed that the downregulation of miR-296-5p or upregulation of RUNX1 could reverse the effects of circ_0000512 knockdown on inhibition of proliferation and promotion of apoptosis in CRC cells. Furthermore, in vivo experiments uncovered that circ_0000512 silence repressed tumor growth through enhancing miR-296-5p and reducing RUNX1. Collectively, these results indicted circ_0000512 exerted its functions via modulating miR-296-5p and RUNX1 expression.
In conclusion, this study illustrated that circ_0000512 was overexpressed in CRC tissues and cells, and circ_0000512 interference inhibited proliferation and facilitated apoptosis by upregulating miR-296-5p and downregulating RUNX1 in CRC cells. In addition, circ_0000512 positively regulated RUNX1 expression by acting as a molecular sponge of miR-296-5p. Collectively, the circ_0000512/miR-296-5p/RUNX1 network may have the potential as a target for the treatment of CRC, which deserves further study and development.
Highlights
The expression of circ_0000512 was enhanced in colorectal cancer tissues and cells.
Knockdown of circ_0000512 inhibited the progression of colorectal cancer by upregulating miR-296-5p and downregulating RUNX1.
Circ_0000512 acted as a molecular sponge of miR-296-5p to modulate RUNX1 expression.
Disclosure
The authors report no funding and no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333. doi:10.1038/nature11928
4. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
5. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
6. Bonizzato A, Gaffo E, Te Kronnie G, Bortoluzzi S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016;6(10):e483. doi:10.1038/bcj.2016.81
7. Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR‐136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247–7256. doi:10.1002/jcp.27482
8. Xu XW, Zheng BA, Hu ZM, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–91683. doi:10.18632/oncotarget.21748
9. Geng Y, Zheng X, Hu W, et al. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci. 2019;133(10):1197–1213. doi:10.1042/CS20190286
10. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi:10.1080/15476286.2016.1220473
11. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-0454-8
12. Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2(4):161–179.
13. Lee KH, Lin FC, Hsu TI, et al. MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim Biophys Acta. 2014;1843(9):2055–2066. doi:10.1016/j.bbamcr.2014.06.001
14. Zhang Z, Zhong X, Xiao Y, Chen C. MicroRNA-296 inhibits colorectal cancer cell growth and enhances apoptosis by targeting ARRB1-mediated AKT activation. Oncol Rep. 2019;41(1):619–629. doi:10.3892/or.2018.6806
15. Ito Y, Bae SC, Chuang LSH. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15(2):81. doi:10.1038/nrc3877
16. Hong D, Fritz AJ, Finstad KH, et al. Suppression of breast cancer stem cells and tumor growth by the RUNX1 transcription factor. Mol Cancer Res. 2018;16(12):1952–1964. doi:10.1158/1541-7786.MCR-18-0135
17. Browne G, Taipaleenmäki H, Bishop NM, et al. Runx1 is associated with breast cancer progression in MMTV‐PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J Cell Physiol. 2015;230(10):2522–2532. doi:10.1002/jcp.24989
18. Ge T, Yin M, Yang M, Liu T, Lou G. MicroRNA-302b suppresses human epithelial ovarian cancer cell growth by targeting RUNX1. Cell Physiol Biochem. 2014;34(6):2209–2220. doi:10.1159/000369664
19. Li Q, Lai Q, He C, et al. RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):334. doi:10.1186/s13046-019-1330-9
20. Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13(2):65–70. doi:10.1016/S0962-8924(02)00043-0
21. Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7(5):812–821. doi:10.1101/gad.7.5.812
22. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
23. Yuan Y, Liu W, Zhang Y, Zhang Y, Sun S. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503(2):870–875. doi:10.1016/j.bbrc.2018.06.089
24. Zhang X, Xu L, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR‐138 targets TERT and PD‐L1. Cell Biol Int. 2017;41(9):1056–1064. doi:10.1002/cbin.10826
25. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
26. Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532–3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505(2):346–352. doi:10.1016/j.bbrc.2018.09.073
27. Fang G, Ye BL, Hu BR, Ruan XJ, Shi YX. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2018;504(1):184–189. doi:10.1016/j.bbrc.2018.08.152
28. Chen Z, Ren R, Wan D, et al. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene. 2019;38(32):6017–6034. doi:10.1038/s41388-019-0857-8
29. Xu C, Li S, Chen T, et al. miR-296-5p suppresses cell viability by directly targeting PLK1 in non-small cell lung cancer. Oncol Rep. 2016;35(1):497–503. doi:10.3892/or.2015.4392
30. Ma X, Zhuang B, Li W. MicroRNA-296-5p downregulated AKT2 to inhibit hepatocellular carcinoma cell proliferation, migration and invasion. Mol Med Rep. 2017;16(2):1565–1572. doi:10.3892/mmr.2017.6701
31. Li T, Lu Y, Zhao X, et al. MicroRNA-296-5p increases proliferation in gastric cancer through repression of Caudal-related homeobox 1. Oncogene. 2014;33(6):783. doi:10.1038/onc.2012.637
32. Li MM, Liu XH, Zhao YC, et al. Long noncoding RNA KCNQ1OT1 promotes apoptosis in neuroblastoma cells by regulating miR‐296‐5p/Bax axis. FEBS J. 2020;287(3):561–577. doi:10.1111/febs.15047
33. He Z, Yu L, Luo S, et al. miR-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4. BMC Cancer. 2017;17(1):140. doi:10.1186/s12885-017-3121-z
34. Felekkis K, Touvana E, Stefanou C, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14(4):236.
35. Zhou Y, Zhang X, Zhang J, Fang J, Ge Z, Li X. LRG1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLoS One. 2017;12(4):e0175122. doi:10.1371/journal.pone.0175122
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.