Back to Journals » OncoTargets and Therapy » Volume 10
Knockdown of a DIS3L2 promoter upstream long noncoding RNA (AC105461.1) enhances colorectal cancer stem cell properties in vitro by down-regulating DIS3L2
Authors Liu W, Yu Q, Ma J, Cheng Y, Zhang H, Luo W, Yao J , Zhang H
Received 18 January 2017
Accepted for publication 6 March 2017
Published 2 May 2017 Volume 2017:10 Pages 2367—2376
DOI https://doi.org/10.2147/OTT.S132708
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Wei Liu,1,* Qiang Yu,2,* Jun Ma,1 Yong Cheng,1 Hongbo Zhang,1 Wengguang Luo,1 Jie Yao,3 Hongyan Zhang1
1Department of Radiation Oncology, Anhui Provincial Hospital, Hefei, 2Department of Hepatobiliary Surgery, 3Cancer Center, Chinese PLA General Hospital & Chinese PLA Medical School, Beijing, People’s Republic of China
*These authors contributed equally to this work
Abstract: A large number of studies have identified plentiful long noncoding RNAs (lncRNAs) associated with the development of multiple cancers. Some lncRNAs have also been found to be strongly linked with stem cell properties such as pluripotency and differentiation. However, only in a few cases have cancer stem cell (CSC)-related lncRNAs been studied. Commonly, the expression and function of lncRNAs are associated with adjacent protein coding transcripts. In the present study, we found an lncRNA (AC105461.1), a promoter upstream transcript of DIS3 mitotic control homolog (Saccharomyces cerevisiae)-like 2 (DIS3L2), may be closely connected with “stem cell-like” properties. We firstly investigated whether the expression of AC105461.1 was down-regulated in colorectal cancer (CRC) tissue samples. Subsequently, we explored the expression pattern of the lncRNA/mRNA gene pair between AC105461.1 and DIS3L2 in 47 CRC specimens by real-time polymerase chain reaction. The results showed that the expression of AC105461.1 was positively correlated with that of DIS3L2. Through CRC cell lines screening experiment, we found that AC105461.1 expression was highest in SW480 and lowest in SW620 cells. Moreover, the results obtained by overexpression experiment indicated that AC105461.1 expression was markedly elevated and DIS3L2 expression level was also apparently upregulated by plasmid cDNA-AC105461.1. In contrast, we further found that AC105461.1 expression level in AC105461.1 siRNA group was significantly knocked down in SW480 cells. Meanwhile, DIS3L2 expression was also markedly decreased. Importantly, we noticed that AC105461.1 overexpression impaired CSC properties, while its knockdown enhanced CSC properties, including self-renewal, migration, and invasion abilities. To further identify the influence of AC105461.1 expression on CSCs properties in CRC, CD133 and CD44, as current universal markers for characterizing CRC stem cells, were selected to perform flow cytometry analysis. As a result, we found that AC105461.1 overexpression reduced the percentage of CD133+CD44+, whereas its knockdown increased the percentage of CD133+CD44+. Taken together, our findings indicated that AC105461.1 may be a regulator of DIS3L2 and a mediator of CRC stem cells, and we speculate that AC105461.1 could be regarded as a promising biomarker and therapeutic target for CRC.
Keywords: AC105461.1, DIS3L2, long noncoding RNA, colorectal cancer, stem cell properties
Introduction
It is well acknowledged that colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide and also associated with high morbidity and mortality.1 Although the treatment for CRC has achieved advances to some extent in recent decades, some patients with CRC often experience recurrence and the clinical results of conventional chemotherapies are still unsatisfactory.2 Currently, with self-renewal and pluripotency as well as differentiation, cancer stem cells (CSCs) are identified as a source of malignant tumors, responsible for tumor aggressiveness and recurrence.3,4 And specially, new therapies targeting CRC cancer stem cells (CRC-CSCs) may markedly raise clinical treatment of CRC to a higher level.5,6
Much work during the past years has been done to reveal that a large portion of the human genome is universally transcribed into noncoding RNAs (ncRNAs). Long noncoding RNAs (lncRNAs) are the RNA molecules larger than 200 nucleotides with no protein-coding capacity.7,8 Recently, lncRNAs expression and their functional mechanisms in tumors have aroused great attention, and numerous cancer-related lncRNAs have been discovered, such as HIF-1 and linc-RoR.9 In addition, some lncRNAs have also been identified to be closely associated with stem cell properties such as pluripotency and differentiation.10,11 Nevertheless, only a few attempts have been made to investigate CSC-related lncRNAs.12
In our previous research work, lncRNA microarray assay was performed on CRC cell lines with the same genetic background but different stem cell properties (data not published). Through bioinformatic analysis, several lncRNAs have been characterized by being connected with CRC-CSC properties. Among them, we explored a potential lncRNA named AC105461.1 (data from UCSC website; http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr2%3A232843265%2D232880417&hgsid=589177787_1PUInTrhbGUiWHLTwt9euHndDLwR). AC105461.1 (ENSG00000227033) starts at 231,978,488 base pair (bp) from pter and ends at 232,015,720 bp from pter, which is located at chr2 (q37.1) on the upstream side of DIS3 mitotic control homolog (Saccharomyces cerevisiae)-like 2 (DIS3L2) and DIS3L2 is also the antisense transcript of it. According to GeneCards (http://www.genecards.org/), both AC105461.1 and DIS3L2 are upregulated in normal colons. Furthermore, they have certain common transcription factors in the corresponding enhancer region, such as EP300, PHF8, POLR2A, and SIN3A.
Generally, the expression and function of lncRNAs are associated with adjacent protein coding transcripts.13,14 Massive lncRNA transcripts emanate from transcription at promoters or other nearby locations of protein-coding genes. As “lncRNA/messenger RNA (mRNA) gene pair”, the interaction between them is associated with stem cell properties.15 DIS3L2 belongs to a family of related 3′–5′ exonucleases with similar domain organization to bacterial RNase II.16 According to GeneCards, one of the functions for DIS3L2 is to mediate the degradation of uridylated pre-let-7 miRNAs, contributing to the maintenance of embryonic stem cells. Intriguingly, a new research demonstrates that DIS3L2 is a new component of the Lin28/let-7 pathway and relevant to Perlman syndrome and cancer.17 Moreover, emerging evidence indicates that Lin28 could promote colon cancer progression and metastasis by upregulating stem cell-related genes or cooperating with WNT signaling.18,19
Therefore, it is tempting to speculate that lncRNA-AC105461.1 may regulate the stem cell-like characteristics of CRC-CSCs by regulating DIS3L2. In the present study, we firstly investigated the expression of AC105461.1 in CRC tissue samples. Next, we explored the expression patterns of the lncRNA/mRNA gene pair between AC105461.1 and DIS3L2 in 47 CRC specimens. Additionally, SW620 and SW480 cells were selected as our candidate cell lines for AC105461.1 overexpression and knockdown experiments. Importantly, cell proliferation assay, matrigel invasion assay, and spheroid formation as well as flow cytometry analysis were performed respectively to investigate the effect of AC105461.1 overexpression or knockdown on stem cell-like properties in vitro.
Materials and methods
Patient samples
Patients with CRC (n=47) who underwent initial surgery in the Chinese People’s Liberation Army (PLA) General Hospital from 2011 to 2012 were selected for this study. Clinical pathology information of all the samples is summarized in Table 1. None of the patients received preoperative therapy such as radiotherapy or chemotherapy prior to surgical resection. The utilization of tumor material for research was approved by the ethical committee of PLA General Hospital. Written informed consent was signed by each participant before tumor sample collection.
  | Table 1 Clinicopathological characteristics of patients with CRC |
Cancer cell lines
Human CRC cell lines SW620, SW480, HT29, HCT116, and Caco-2 derived from normal human colon were all acquired from the American Type Culture Collection (ATCC; Manassas, VA, USA). The SW620 and SW480 cells were cultured in Leibovitz’s L-15 Medium (Thermo Fisher Scientific, Waltham, MA, USA) and the other CRC cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) medium (Thermo Fisher Scientific). All these media contained 10% fetal bovine serum (Thermo Fisher Scientific) and were maintained at 37°C with 5% CO2. CSCs in SW620 and SW480 were enriched for CSC-related experiments. The SW620 and SW480 cells were transferred into the stem cell-culturing medium, serum-free DMEM/F12 medium, which consists of 20 ng/mL human epidermal growth factor (Peprotech Inc, Rocky Hill, NJ, USA), 20 ng/mL human basic fibroblast growth factor (Peprotech Inc), and 1% N2 supplement (Thermo Fisher Scientific).
Quantitative real-time polymerase chain reaction (PCR)
Total RNA was extracted from CRC tumor tissues and matched adjacent normal tissues, using Trizol Total RNA Reagent (Thermo Fisher Scientific) following the manufacturer’s protocol. The first strand complementary DNAs (cDNAs) were synthesized with 2 μg total RNAs by means of RevertAidTM H Minus First Strand cDNA Synthesis Kit (Takara, Otsu, Japan). The primer sequences were designed and synthesized by GenePharma (Shanghai, People’s Republic of China). Quantitative real-time PCR was performed using the SYBR PrimeScript RT-PCR kit (Takara) using Applied Biosystems 7500 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s recommendations. The reaction conditions were designed as follows: 1 cycle at 95°C for 30 seconds followed by 40 cycles at 95°C for 5 seconds, and 60°C for 34 seconds. The ΔΔCT calculation with CT served as the threshold cycle for quantification of gene expression, and the target gene expression level in patients was measured as “ratio (target in tumor tissue/target in nontumorous tissue”: R (T/N)). All reactions were performed in triplicate and normalized by the internal control products of GAPDH. All sequences we used are listed in Table 2.
  | Table 2 Primers for real-time PCR analysis |
Transfection of small interfering RNA (siRNA)
For siRNA analysis, siRNA for the AC105461.1 sequence and non-targeting siRNA were obtained from GenePharma. The sequences are listed in Table 3. Prior to transfection, approximately 5% SW620 and SW480 cells were plated into 12-well plates and cultured for at least 24 hours to achieve a confluence of 30%–50%. SiRNA transfection was conducted using X-treme GENE™ transfection reagent (Hoffman-La Roche Ltd, Basel, Switzerland) following the manufacturer’s instructions. After transfection, cells were harvested for cell proliferation assay, matrigel invasion assay, and spheroid formation assay.
  | Table 3 Sequences for small interfering RNA analysis |
Overexpression of AC105461.1 in CRC cells
The AC105461.1 low-expressed SW620 cells were selected to be transfected with plasmid complementary DNA (pcDNA)-AC105461.1 by Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. AC105461.1 cDNA was cloned into BamHI-EcoRI sites of pcDNA3.1. Those transfected cells were harvested for RNA isolation, cell proliferation assay, matrigel invasion assay, spheroid formation assay, and flow cytometry analysis.
Cell proliferation assay
According to the manufacturer’s instruction, cell proliferation ability was measured by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega, Madison, WI, USA) after transfection. The SW620 and SW480 cells were plated separately in 96-well at a density of 2,000 cells per well after transfection. Each well containing 100 μL culture medium was added 20 μL of the MTS reagent. The plate was incubated for 2 hours at 37°C in a humidified, 5% CO2 atmosphere. Afterward, the plate was read at 490 nm using a plate reader.
Matrigel invasion assay
Transwell assay was performed to observe the cells’ invasion ability, and it was conducted using modified Boyden Chambers, which consisted of transwell-precoated matrigel membrane filter with 8 mm pores inserting in 24-well tissue culture plates (BD Biosciences, San Jose, CA, USA). DMEM supplemented with 10% inactivated fetal bovine serum was used as chemoattractant in the lower chamber. Cells that had migrated through the filter were stained and counted at regular intervals. The mean migration rate was calculated as the increasing radius of the entire cell population over time.
Spheroid formation assay
Based on ultralow attachment surface 96-well culture plates (Corning Incorporated, Corning, NY, USA), spheroid formation assay was carried out to evaluate the capability of cell self-renewal. Cells in the pcDNA-AC105461.1 and pcDNA control, or in the AC105461.1 siRNAs and negative control siRNA (NC-si) were respectively resuspended in 200 μL serum-free medium DMEM/F12 at a density of 200 cells per well. DMEM/F12 was supplemented with 20 ng/mL human epidermal growth factor, 20 ng/mL human basic fibroblast growth factor, and 1% N2. Phase-contrast images were conducted 7 days later.
Flow cytometry analysis
Cells prepared for analyzing the cell surface markers were plated at 60% confluence the day before analysis. Trypsinized cells were washed by phosphate buffered saline, resuspended in 1% bovine serum albumin plus corresponding fluorophore-conjugated primary antibodies, and then incubated for 30 minutes at room temperature. Afterward, the expression of CD133 and CD44 in cultured cells was analyzed by flow cytometry (BD Biotechnology). CD44 and CD133 in SW480 or SW620 cells were respectively sorted by fluorescence-activated cell sorter to obtain CD133+CD44+, CD133+CD44−, CD133−CD44+, and CD133−CD44− subpopulations.
Statistical analyses
Each mentioned experiment was performed at least three times. Differences between groups were analyzed using the Student’s t-test. Correlation between gene expressions was compared through Pearson’s correlation. Statistical analyses were performed using SPSS version 18.0 (SPSS Inc, Chicago, IL, USA). All analyses were considered statistically significant at P<0.05(*).
Results
The expression of AC105461.1 was down-regulated in CRC tissue samples
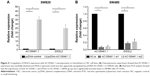
The expression of AC105461.1 was assessed by a panel paired specimen obtained from 47 CRC patients. The results indicated that the expression of AC105461.1 was significantly down-regulated in CRC tissue samples (Figure 1, *P<0.05).
  | Figure 1 The expression of AC105461.1 was down-regulated in CRC tissue samples (*P<0.05). |
AC105461.1 expression was correlated with DIS3L2 in CRC tissue samples
To identify the relationship between AC105461.1 and DIS3L2, we assessed the correlation of their expression levels in a group of 47 CRC patients using real-time PCR. For every sample, lncRNA was separated from cancerous tissues and adjacent nontumorous CRC tissues. The results illustrated that the expression of AC105461.1 was correlated with that of DIS3L2 in CRC tissue samples (Figure 2, R=0.485, P<0.001).
  | Figure 2 AC105461.1 expression was correlated with DIS3L2 (P<0.001). |
The expression of AC105461.1 was assessed in different CRC cell lines
Through real-time PCR, we explored AC105461.1 expression in five CRC cell lines (SW480, HT29, HCT116, Caco-2, and SW620). The results demonstrated that the expression of AC105461.1 was highest in SW480 and lowest in SW620 cells (Figure 3, *P<0.05). Hence, SW620 and SW480 cells were respectively selected as our candidate cell lines for AC105461.1 overexpression and knockdown experiments.
  | Figure 3 AC105461.1 expression levels were assessed by real-time PCR in five cell lines (*P<0.05). |
Coregulation of DIS3L2 expression with AC105461.1 overexpression or knockdown in CRC cell lines
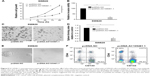
In the overexpression experiment, pcDNA-AC105461.1 and pcDNA control were conducted and transfected into SW620 cells. The results indicated that AC105461.1 expression was markedly elevated and DIS3L2 expression level was also apparently upregulated by pcDNA-AC105461.1 (Figure 4A, *P<0.05).
In the knockdown experiment, AC105461.1 siRNA and NC-si were respectively transfected into SW480 cells. Quantification PCR analysis showed that AC105461.1 expression level in AC105461.1 siRNA group was significantly knocked down in SW480 cells. Similarly, DIS3L2 expression level was also markedly decreased (Figure 4B, *P<0.05). Consequently, we could roughly speculate that the DIS3L2 expression may be modulated by AC105461.1.
AC105461.1 overexpression impaired the CSC properties
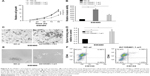
In this section, the effect of AC105461.1 overexpression on stem cell-like properties in vitro was investigated in SW620 cells. Cell proliferation assay was conducted by transfecting pcDNA-AC105461.1 and NC plasmid into SW620 cells. The results showed that cell growth of SW620 cells in pcDNA-AC105461.1 was evidently slower than that in pcDNA-NC (Figure 5A, *P<0.05). The matrigel invasion assay conducted in SW620 cells exerted significant cell invasion inhibition in pcDNA-AC105461.1 compared with pcDNA-NC (Figure 5B and C, *P<0.05). Additionally, spheroid formation assay revealed that the spheroid formation rate in pcDNA-AC105461.1 was obviously slower compared with that in pcDNA-NC (Figure 5D and E, *P<0.05). Furthermore, flow cytometric analysis was conducted to further identify the influence of AC105461.1 overexpression on the CRC-CSCs properties. CD133 and CD44 were selected as the surface markers of CRC in our study. We found that the percentage of CD133+CD44+ in pcDNA-AC105461.1 group (2.01%) was low when compared with pcDNA-NC group (12.10%) (Figure 5F). Taken together, these results suggested that AC105461.1 overexpression could impair the CSC properties.
AC105461.1 knockdown enhanced the CSC properties
In this section, we determined the effect of AC105461.1 knockdown on stem cell-like properties in vitro in SW480 cells. Cell proliferation assay was conducted by transfecting AC105461.1 siRNA group and NC-si into SW480 cells. The results illustrated that cell growth of SW480 cells in AC105461.1 siRNA group was markedly higher than that in NC-si (Figure 6A, *P<0.05). Meantime, the matrigel invasion assay also showed significant cell invasion enhancement in AC105461.1 siRNA group in contrast to the NC group in SW480 cells (Figure 6B and C, *P<0.05). In addition, spheroid formation assay demonstrated that the spheroid formation rate in AC105461.1-si1 was obviously higher than that in NC-si (Figure 6D and E, *P<0.05). Flow cytometric analysis was also performed to observe the impact of AC105461.1 knockdown on CRC-CSCs properties. We observed that the percentage of CD133+CD44+ in AC105461.1 siRNA group was 4.39% and that in NC-si group was 0.96%, which illustrated that AC105461.1 knockdown enhanced the percentage of CD133+CD44+ (Figure 6F). These results indicated that AC105461.1 knockdown could enhance the CSC properties.
Discussion
Recent studies have proved that the lncRNAs have an impact on cancer development.20,21 LncRNAs expression and their functional mechanisms have aroused great attention in cancer diagnosis and treatment.22 Emerging studies identify that some well-known cancer-related lncRNAs, such as lncRNA-ATB,23 lncRNA-CLMAT3,24 and lncRNA-H19,25 are also involved in the tumor progression of CRC.
AC105461.1 is the antisense transcript of DIS3L2. A new study has demonstrated that DIS3L2 is a new component of the Lin28/let-7 pathway and also relevant to Perlman syndrome and cancer.17 Additionally, King et al18 and Tu et al19 reported that Lin28 could promote colon cancer progression and metastasis by upregulating stem cell-related genes or cooperating with WNT signaling. Moreover, it has been revealed that transcriptions of most lncRNA genes are coordinated with that of adjacent protein-coding genes.15 Intriguingly, AC105461.1 and DIS3L2 could form “lncRNA/mRNA gene pair”, which have impact on each other in expression and function and also are closely associated with stem cell properties.15
In the present study, we firstly investigated whether the expression of AC105461.1 was down-regulated in CRC tissue samples. Next, we explored the expression patterns of the lncRNA/mRNA gene pair between AC105461.1 and DIS3L2 in 47 CRC specimens by real-time PCR. The results showed that the expression of AC105461.1 was positively correlated with that of DIS3L2 in CRC tissue samples. In order to select the appropriate cell lines, we investigated the expression of AC105461.1 in five different CRC cell lines. SW620 and SW480 cells were selected as our candidate cell lines for AC105461.1 overexpression and knockdown experiment, respectively. According to the AC105461.1 overexpression experiment, we found that AC105461.1 expression was markedly elevated in SW620 cells and DIS3L2 expression level was also apparently upregulated. Meantime, the results obtained by the AC105461.1 knockdown experiment suggested that AC105461.1 expression level in AC105461.1 siRNA group was significantly knocked down in SW480 cells and DIS3L2 expression was also markedly decreased. These results made us speculate that the DIS3L2 expression may be modulated by AC105461.1.
Currently, only a small number of CSC-like properties of CSC-related lncRNAs have been investigated.12 Herein, cell proliferation assay, matrigel invasion assays, and spheroid formation as well as flow cytometry analysis were implemented to analyze the effect of AC105461.1 overexpression or knockdown on stem cell-like properties in vitro. We found that AC105461.1 overexpression impaired the CSC properties while knockdown enhanced the CSC properties, including self-renewal, migration, and invasion abilities. To further identify the influence of AC105461.1 expression on CRC-CSCs properties, CD133 and CD44 markers were selected to perform the flow cytometry analysis. The results indicated that overexpressed AC105461.1 reduced the percentage of CD133+CD44+ while knockdown showed the opposite effect. To our knowledge, this study may provide the first evidence that AC105461.1 is likely to negatively correlate with tumor progression. In addition, further studies declare that DIS3L2 is strongly linked with Lin28/let-7 pathway which is relevant to various cancers.17,26,27 The need for further analyses of the exact regulation mechanism and the relationship between AC105461.1 and Lin28/let-7 pathway are unmet. We will perform the relevant experiments in our future study.
Conclusion
Collectively, our data suggest that AC105461.1 may serve as a regulator of DIS3L2 and a mediator of CSCs in CRC. Furthermore, it could be a potential biomarker and therapeutic target for CRC. Certainly, further work targeting the exact regulation mechanism and the relationship between AC105461.1 and Lin28/let-7 pathway in CRC will be of great significance in the clinical cancer research.
Disclosure
The authors report no conflicts of interest in this work.
References
Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–197. | ||
Garcia-Foncillas J, Diaz-Rubio E. Process in metastatic colorectal cancer: growing role of cetuximab to optimize clinical outcome. Clin Transl Oncol. 2010;12(8):533–542. | ||
Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5 (+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526(7575):715–718. | ||
Shibata M, Shen MM. The roots of cancer: stem cells and the basis for tumor heterogeneity. Bioessays. 2013;35(3):253–260. | ||
De Angelis ML, Zeuner A, Policicchio E, et al. Cancer stem cell-based models of colorectal cancer reveal molecular determinants of therapy resistance. Stem Cells Transl Med. 2016;5(4):511–523. | ||
Fanali C, Lucchetti D, Farina M, et al. Cancer stem cells in colorectal cancer from pathogenesis to therapy: controversies and perspectives. World J Gastroenterol. 2014;20(4):923–942. | ||
Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. | ||
Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24(4):616–628. | ||
Chang YN, Zhang K, Hu ZM, et al. Hypoxia-regulated lncRNAs in cancer. Gene. 2016;575(1):1–8. | ||
Wang F, Yuan JH, Wang SB, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60(4):1278–1290. | ||
Fanale D, Barraco N, Listì A, Bazan V, Russo A. Non-coding RNAs functioning in colorectal cancer stem cells. Adv Exp Med Biol. 2016;937:93–108. | ||
Wang X, Sun W, Shen WF, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016; 64(6):1283–1294. | ||
Preker P, Almvig K, Christensen MS, et al. Promoter upstream transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39(16):7179–7193. | ||
Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle. 2009;8(15):2332–2338. | ||
Sigova AA, Mullen AC, Molinie B, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110(8):2876–2881. | ||
Astuti D, Morris MR, Cooper WN, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44(3):277–284. | ||
Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497(7448):244–248. | ||
King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011;71(12):4260–4268. | ||
Tu HC, Schwitalla S, Qian Z, et al. LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev. 2015;29(10):1074–1086. | ||
Zhang YH, Fu J, Zhang ZJ, Ge CC, Yi Y. LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8(12):5286–5297. | ||
Ye C, Shen Z, Wang B, et al. A novel long non-coding RNA lnc-GNAT1-1 is low expressed in colorectal cancer and acts as a tumor suppressor through regulating RKIP-NF-κB-Snail circuit. J Exp Clin Cancer Res. 2016;35(1):187. | ||
Sültmann H, Diederichs S. Long noncoding RNA: “LNCs” to cancer. Eur Urol. 2014;65(6):1152–1153. | ||
Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35(3):1385–1388. | ||
Ye LC, Ren L, Qiu JJ, et al. Aberrant expression of long noncoding RNAs in colorectal cancer with liver metastasis. Tumour Biol. 2015;36(11):8747–8754. | ||
Han D, Gao X, Wang M, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7(16):22159–22173. | ||
Faehnle CR, Walleshauser J, Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014;514(7521):252–256. | ||
Suzuki HI, Katsura A, Miyazono K. A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B. Cancer Sci. 2015;106(9):1174–1181. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.