Back to Journals » Cancer Management and Research » Volume 10
KLF6-SV1 is a new prognostic biomarker in postoperative patients with non-small cell lung cancer
Authors Zhang N, Li Z, Xiao W, Yang F, Gao W, Sun ZG
Received 20 April 2018
Accepted for publication 19 July 2018
Published 26 September 2018 Volume 2018:10 Pages 3937—3944
DOI https://doi.org/10.2147/CMAR.S171805
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Nan Zhang,1 Zhe Li,2 Wei Xiao,3 Fei Yang,4 Wei Gao,4 Zhi-Gang Sun3
1Department of Oncology, Jinan Central Hospital Affiliated to Shandong University, Jinan, People’s Republic of China; 2Department of Medical Examination, Jinan Central Hospital Affiliated to Shandong University, Shandong University, Jinan, People’s Republic of China; 3Department of Thoracic Surgery, Jinan Central Hospital Affiliated to Shandong University, Shandong University, Jinan, People’s Republic of China; 4Department of Pathology, Jinan Central Hospital Affiliated to Shandong University, Shandong University, Jinan, People’s Republic of China
Objectives: Non-small cell lung cancer (NSCLC) is aggressive and associated with a poor prognosis. Recent studies have revealed that several genes are involved in the origin and progression of NSCLC. Kruppel-like factor 6 (KLF6) inactivation has been shown in some malignant tumors. KLF6-SV1, as one of the alternatively spliced KLF6 isoforms, has been found to be correlated with metastatic potential and poor survival in some cancers. The purpose of this study was to investigate the clinical and prognostic significance of KLF6-SV1 expression in NSCLC patients after curative resection.
Patients and methods: A total of 79 patients were enrolled in this study. Enumeration data were analyzed using the chi-squared test or Fisher’s exact probability test. Measurement data were represented as average±SD and t-test (homoscedasticity) or t’-test (homoscedasticity uneven). Univariate analysis was performed by modeling Kaplan–Meier survival curves. The log-rank test was used to calculate the survival rate. Multivariate analysis was carried out by the use of the Cox proportional hazard model.
Results: KLF6-SV1 expression was correlated with pN (P<0.05) and pTNM stage (P<0.05). The expression of KLF6-SV1 in the adenocarcinoma group was significantly higher than that in the squamous cell carcinoma group (P<0.05). The 5-year survival rate for 79 NSCLC patients was 40.5%, and it was significantly associated with differentiation (P<0.05), pN (P<0.01), pTNM stage (P<0.01) and high expression of KLF6-SV1 (P<0.01). Cox multivariate regression demonstrated that differentiation, pN and KLF6-SV1 expression were independent factors for the 5-year survival rate.
Conclusion: KLF6-SV1 expression in adenocarcinoma was significantly higher than that in the squamous cell carcinoma, and high expression of KLF6-SV1 was significantly associated with pN and pTNM stage and poor survival in NSCLC patients.
Keywords: KLF6-SV1, non-small cell lung cancer, real-time PCR, Western blot, immunohistochemistry
Introduction
Lung cancer is the most common cause of tumor-related mortality in the world, especially in China. More than 85% of lung neoplasm is non-small cell lung cancer (NSCLC), and squamous cell carcinoma and adenocarcinoma are still the main subtypes.1 So far, for patients with stage I–IIIa NSCLC, surgery might be the most effective treatment. However, about 75% of the NSCLC patients have advanced disease at their first visit.2 The prognosis of the NSCLC patients is unsatisfactory, and the 5-year survival rate is about 15%.3 The TNM staging system4 is short of enough predictive value because significant differences in survival are often found in the identical TNM stage. Therefore, it is of great value to combine some biomarkers to distinguish NSCLC patients with poor survival.5,6 Kruppel-like factor 6 (KLF6) (GeneBank accession number AF001461), a member of the KLF family, is a group of zinc finger transcription factors that are associated with regulating cellular differentiation, development, proliferation, growth-associated signal transduction and apoptosis.7,8 KLF6 inactivation has been shown in some malignant tumors. Three alternatively spliced KLF6 isoforms have been found in a previous study.9 Among them, KLF6-SV1 has been shown to promote tumor growth by antagonizing the tumor suppressor function of KLF6.10,11 High expression of KLF6-SV1 in some cancer cell lines could lead to tumor invasion and metastasis.12,13
A very limited number of studies have shown the clinical features of KLF6-SV1 in NSCLC patients. The purpose of this study was to investigate the association between KLF6-SV1 expression and the clinicopathological features of NSCLC patients. KLF6-SV1 expression was detected by real-time PCR at the mRNA level and by Western blot and immunohistochemistry at the protein level. In addition, the study also investigated the potential prognostic value of KLF6-SV1 to predict the NSCLC patients’ survival rate by the univariate and multivariate analyses.
Patients and methods
Patients
A total of 79 patients were enrolled in this study from the Department of Thoracic Surgery, Jinan Central Hospital, between January 2009 and December 2012. The inclusion criteria were as follows: 1) patients accepted radical operation and affirmed squamous cell carcinoma or adenocarcinoma by pathology; 2) patients were diagnosed with stage I–IIIa NSCLC, according to the TNM staging system summarized by the International Union Against Cancer (2009)4; 3) patients accepted no presurgical chemotherapy or radiotherapy; and 4) patients had no seriously surgical contraindications. Large cell carcinoma and adenosquamous carcinoma were excluded because of too small sample sizes. Tables 1 and 2 show the clinicopathological features of the patients.
Tissue samples
We obtained NSCLC specimens from the 79 patients. Ten corresponding normal lung tissue samples that were randomly sampled from 10 of the 79 patients’ paracancerous normal lung tissues (at least 2 cm away from cancer) were used as controls. Each specimen was divided into two parts. A minimal of 0.4 cm×0.4 cm×0.4 cm tissue specimen was wrapped in a foil quickly after being labeled and then snap frozen in liquid nitrogen for 1 minute and kept at −80°C for real-time PCR and Western blot analysis. The other tissue specimen was fixed in formalin and then embedded in paraffin for histopathological examination and immunohistochemistry.
Real-time PCR
The total RNA isolated from sample specimens was performed according to a previous study.14 The first strand cDNA was synthesized using PrimeScript™ RT Master Mix (TaKaRa, Dalian, China). The sense and antisense primers were synthesized as follows: actin forward, 5′-CTGAAGTACCCCATCGAGCAC-3′ and reverse, 5′-ATAGCACAGCCTGGATAGCAAC-3′; KLF6-SV1 forward, 5′-GACCAAAATCATTCTGGCTCG-3′ and reverse, 5′-GATTCGCTGCTGACATCTGAGT-3′. Quantitative real-time PCR was carried out on an ABI 7500 PCR (Thermo Fisher Scientific, Waltham, MA, USA) system under the following conditions: an initial denaturation at 95°C for 30 seconds, followed by (95°C for 5 seconds and 60°C for 40 seconds)×40 cycles for the target gene. The fold change in gene expression was evaluated by the 2–ΔΔCt method.
Western blot
Western blot was performed according to the method described in a previous study.15 In brief, the micrograms of the proteins from each sample were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk, incubated with primary mouse antibody against human KLF6-SV1 (1:500, Product 39-6900; Thermo Fisher Scientific) and mouse monoclonal β-actin antibody (1:5,000, Product TA-09; Zhongshanjinqiao, Beijing, China) overnight at 4°C and finally with Peroxidase-Conjugated Goat anti-Mouse IgG (1:5,000, Product ZB-2305; Zhongshanjinqiao). Immunoblotted proteins were visualized by enhanced chemiluminescence reagents, and the signals were detected by Alphaimager 2200 imaging system (Alphaimager, San Francisco, CA, USA) and Image J analysis software. The formula used to calculate the relative KLF6-SV1 expression was as follows: gray value (KLF6-SV1)/gray value (β-actin).
Immunohistochemistry
Immunohistochemistry staining for KLF6-SV1 was detected by the streptavidin–peroxidase (SP) method.16 In brief, sample sections were incubated overnight at 4°C with primary mouse antibody against human KLF6-SV1 (1:120, Product 39-6900; Thermo Fisher Scientific). A secondary antibody was then added using the horseradish peroxidase (HRP)-Sp9002 System (SPlink Detection Kits, Biotin-Streptavidin HRP Detection Systems; Zhongshanjinqiao) according to the manufacturer’s instructions. KLF6-SV1 expression levels were measured using a semiquantitative immunoreactivity scoring system (IRS) as described previously.16 The cases were grouped as low expression (IRS 0–6) and high expression (IRS 7–12), respectively. Five randomly chosen microscopic fields were examined at high magnification (200×) under a light microscope.
Statistical analyses
Enumeration data were analyzed using the chi-squared test or Fisher’s exact probability test. Measurement data were represented as average±SD and t-test (homoscedasticity) or t′-test (homoscedasticity uneven). Univariate analysis was performed by modeling Kaplan–Meier survival curves. The log-rank test was used to calculate the survival rate. Multivariate analysis was carried out by the use of the Cox proportional hazard model. All statistical data were analyzed using SPSS (version 13; SPSS Inc, Chicago, IL, USA), and P<0.05 indicates a statistically significant difference.
Follow-up
In total, 61 patients received postsurgical chemotherapy, 31 patients received postsurgical radiotherapy and 28 patients received epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) therapy. The location and time of tumor relapse were recorded. Patients who succumbed to mortality due to the tumor were included in the prognostic analysis.
Ethics statement
The study was approved by Jinan Central Hospital Affiliated to Shandong University (Jinan, People’s Republic of China). Written informed consent was obtained from all the 79 patients.
Results
Real-time-PCR
KLF6-SV1 mRNA expression was detected in all the patients by the real-time PCR. The expression of KLF6-SV1 mRNA in the tumor region was significantly higher than that in the paracancerous region corresponding to the normal lung tissues (t, t′ 3.970; P=0.001) (Table 3). As shown in Table 1, KLF6-SV1 mRNA expression was correlated with pN (t, t′ 5.136, P=0.001), pTNM stage (P<0.05) (pI vs pII, t, t′ 4.280, P=0.001; pI vs pIIIa, t, t′ 6.416, P=0.001; pII vs pIIIa, t, t′ 2.219, P=0.030) and the 5-year survival rate (t, t′ 9.054, P=0.001). Moreover, the expression of KLF6-SV1 in the adenocarcinoma group was significantly higher than that in the squamous cell carcinoma group (t, t′ 2.156; P=0.034).
Western blot
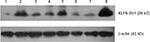
KLF6-SV1 protein expression was detected in all the patients by Western blot. The expression of KLF6-SV1 protein in the tumor region was significantly higher than that in the paracancerous region corresponding to normal lung tissues (t, t′ 3.978, P=0.001) (Table 3). As shown in Table 1, KLF6-SV1 protein expression was correlated with pN (t, t′ 5.204, P=0.001), pTNM stage (P<0.05) (pI vs pII, t, t′ 4.290, P=0.001; pI vs pIIIa, t, t′ 6.626, P=0.001; pII vs pIIIa, t, t′ 2.285, P=0.026) and the 5-year survival rate (t, t′ 9.372, P=0.001). Furthermore, the protein expression of KLF6-SV1 in the adenocarcinoma group was significantly higher than that in the squamous cell carcinoma group (t, t′ 2.206, P=0.046) (Figure 1).
Immunohistochemistry
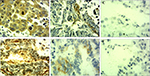
KLF6-SV1 protein expression in all the patients was also detected by immunohistochemistry. The positive KLF6-SV1 protein expression was mainly located in the cytoplasm (Figure 2). Moreover, the high protein expression of KLF6-SV1 in the tumor region was significantly higher than that in the paracancerous region corresponding to normal lung tissues (57.0% vs 0.0%, P=0.001) (Table 3). Table 2 demonstrates that the high expression of KLF6-SV1 was significantly associated with pathological type (squamous cell carcinoma 45.2% vs adenocarcinoma 70.3%; P=0.025), pathological lymph node (pN– 39.3% vs pN+ 66.7%; P=0.019) and pTNM stage (pI, 31.6% vs pII, 60.5% vs pIIIa, 76.5%; P<0.05). The 5-year survival rate for the 79 NSCLC patients was 40.5%. A univariate analysis was conducted using the log-rank test, and the 5-year survival rate was significantly associated with differentiation (P<0.05), pN (P=0.001), pTNM stage (P<0.01) and high expression of KLF6-SV1 (P=0.001) (Figure 3 and Table 4). Cox multivariate regression demonstrated that differentiation, pN and KLF6-SV1 expression were independent factors for the 5-year survival rate (Table 5).
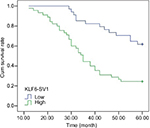  | Figure 3 A Kaplan–Meier analysis of the cumulative survival rate after operation in patients with high and low expression of KLF6-SV1, respectively. Abbreviation: KLF6, Kruppel-like factor 6. |
Discussion
Up to now, KLF6-SV1, KLF6-SV2 and KLF6-SV3 have been identified as three alternatively spliced KLF6 isoforms.17 Debouki-Joudi et al18 used quantitative reverse transcription PCR to detect wild-type transcript KLF6 (wtKLF6) and the spliced variants (KLF6-SV1, KLF6-SV2 and KLF6-SV3) at the mRNA level in a total of 50 nasopharyngeal carcinoma patients, and they demonstrated that the level of wtKLF6 was significantly lower in tumors than in normal tissues (P=0.0015). The spliced variants KLF6-SV1 and KLF6-SV2 were significantly overexpressed than the wtKLF6 in tumor cases (P<0.0001 and P=0.02215, respectively), whereas no difference was seen between wtKLF6 and KLF6-SV3 (P=0.4928). Moreover, they also found that KLF6-SV1 was significantly higher in young patients than in adult patients (P=0.0003). Zhang et al19 used qRT-PCR to detect wtKLF6, KLF6-SV1, KLF6-SV2 and KLF6-SV3 at the mRNA level in a total of 60 colorectal cancer (CRC) patients and found that the splice variants represented 1.5%–15.9% compared with wt KLF6, and SV1 mRNA expression was higher than SV2 and SV3 mRNA expression. The total, wild type, SV2 and SV3 of KLF6 were decreased except for SV1 expression increased in CRC samples. These studies show that KLF6-SV1 takes an important function in some cancers. KLF6-SV1 is also observed to be overexpressed in head and neck cancers and breast malignant tumor and was correlated with metastatic potential and with poor survival.20,21 Furthermore, in ovarian and prostate cancer models, KLF6-SV1 is found to antagonize tumor suppressor function of KLF6 and can promote tumor growth and progression.12,22 It is also found that in vitro, siRNA-mediated downregulation of KLF6-SV1 could decrease the tumor growth. The role of KLF6-SV1 in lung cancer remains largely unknown. Only two papers have been found in PubMed to show the role of KLF6-SV1 in lung adenocarcinoma. DiFeo et al23 used PCR to detect KLF6-SV1 expression at the mRNA level in a total of 70 lung adenocarcinoma patients and they demonstrated that KLF6-SV1 mRNA was overexpressed in lung adenocarcinoma and was correlated with poor survival in adenocarcinoma patients. Sangodkar et al24 reported that high KLF6-SV1expression was found in chemoresistant adenocarcinoma cancer cells in vitro both at mRNA and protein levels. Through induction of apoptosis, siRNA-mediated downregulation of KLF6-SV1 could restore chemotherapy sensitivity to lung adenocarcinoma cells. Their data also showed that KLF6-SV1 overexpression could lead to reduced chemotherapy sensitivity in lung adenocarcinoma cells.
A total of 79 NSCLC patients were enrolled in this study. All the patients underwent complete resection. The KLF6-SV1 expression was detected both by real-time PCR at the mRNA level and Western blot and immunohistochemistry at the protein level. Moreover, KLF6-SV1 expression was quantified by both qualitative and quantitative analyses at the protein level. Our results showed that high expression of KLF6-SV1 in the tumor region was significantly higher than that in the paracancerous region corresponding to normal lung tissues. The high expression of KLF6-SV1 was correlated with pN and pTNM stage. The expression of KLF6-SV1 in the adenocarcinoma group was significantly higher than that in the squamous cell carcinoma group. Our data showed that the 5-year survival rate of NSCLC patients was 40.5%, and it was significantly associated with the degree of differentiation, pN, pTNM stage and the KLF6-SV1 expression. To eliminate the impact of mixed factors on statistical analysis, we used multivariate analysis to determine prognostic factors, and the result showed that pN and high expression of KLF6-SV1 were relevant independent factors for a poor prognosis. Our data demonstrated that the high expression of KLF6-SV1 was correlated with metastatic potential and with poor survival in the NSCLC patients.
In China, the indications for treatment not only depend on doctors’ preferences but also on patients’ willingness and economic status. In this study, 61 patients received postoperative chemotherapy, 31 patients received postoperative radiotherapy and 28 patients received EGFR-TKI therapy. However, neither univariate nor multivariate analysis showed statistically significant correlations with postoperative chemotherapy, radiotherapy and EGFR-TKI therapy on the 5-year survival rate. This study is based on the limited number of patients from our hospital, and we will further validate our conclusion by exploring more patients at multiple hospitals.
Conclusion
KLF6-SV1 expression in adenocarcinoma was significantly higher than that in the squamous cell carcinoma, and the high expression of KLF6-SV1 was significantly associated with pN and pTNM stage and poor survival in NSCLC patients. Our data suggest that KLF6-SV1 can potentially be a good biomarker for NSCLC, and the targeted reduction of KLF6-SV1 might represent a novel therapeutic strategy for NSCLC treatment.
Acknowledgments
This study was funded by the grants from the National Natural Science Foundation of China (No 81502525) and the Second Group of Jinan Science and Technology Development Program (No 201602204).
Disclosure
The authors report no conflicts of interest in this work.
References
Li Y, Wei S, Wang J, Hong L, Cui L, Wang C. Analysis of the factors associated with abnormal coagulation and prognosis in patients with non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2014;17(11):789–796. | ||
Sun W, Song L, Ai T, Zhang Y, Gao Y, Cui J. Prognostic value of MET, cyclin D1 and MET gene copy number in non-small cell lung cancer. J Biomed Res. 2013;27(3):220–230. | ||
Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. | ||
Mirsadraee S, Oswal D, Alizadeh Y, Caulo A, van Beek E. The 7th lung cancer TNM classification and staging system: review of the changes and implications. World J Radiol. 2012;4(4):128–134. | ||
Tan Z, Yang C, Zhang X, Zheng P, Shen W. Expression of glucose transporter 1 and prognosis in non-small cell lung cancer: a pooled analysis of 1665 patients. Oncotarget. 2017;8(37):60954–60961. | ||
Zhou J, Yu Y, Pei Y, et al. A potential prognostic biomarker SPC24 promotes tumorigenesis and metastasis in lung cancer. Oncotarget. 2017;8(39):65469–65480. | ||
Wen PH, Wang DY, Zhang JK, et al. Kruppel-like factor 6 suppresses growth and invasion of hepatocellular carcinoma cells in vitro and in vivo. Int J Immunopathol Pharmacol. 2016;29(4):666–675. | ||
Narla G, Friedman SL, Martignetti JA. Krüppel cripples prostate cancer: KLF6 progress and prospects. Am J Pathol. 2003;162(4):1047–1052. | ||
Difeo A, Martignetti JA, Narla G. The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat. 2009;12(1-2):1–7. | ||
Tchirkov A, Sapin V, Marceau G, et al. Increased expression of the oncogenic KLF6-SV1 transcript in human glioblastoma. Clin Chem Lab Med. 2010;48(8):1167–1170. | ||
Narla G, Difeo A, Fernandez Y, et al. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J Clin Invest. 2008;118(8):2711–2721. | ||
Narla G, Difeo A, Yao S, et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005;65(13):5761–5768. | ||
Chen H, Chen L, Sun L, Zhen H, Li X, Zhang Q. A small interfering RNA targeting the KLF6 splice variant, KLF6-SV1, as gene therapy for gastric cancer. Gastric Cancer. 2011;14(4):339–352. | ||
Cao Y, Song J, Chen J, Xiao J, Ni J, Wu C. Overexpression of NEK3 is associated with poor prognosis in patients with gastric cancer. Medicine. 2018;97(3):e9630. | ||
Hu B, Jiang D, Chen Y, et al. High CHMP4B expression is associated with accelerated cell proliferation and resistance to doxorubicin in hepatocellular carcinoma. Tumour Biol. 2015;36(4):2569–2581. | ||
Zhang Q, Tan XP, Yuan YS, et al. Decreased expression of KLF6 and its significance in gastric carcinoma. Med Oncol. 2010;27(4):1295–1302. | ||
Narla G, Difeo A, Reeves HL, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65(4):1213–1222. | ||
Debouki-Joudi S, Mhirsi S, Mokni-Baizig N, et al. Overexpression of the oncogenic variant (KLF6-SV1) in young NPC patients and correlation with lack of e-csadherin. Anal Cell Pathol. 2018;2018:9654067–7. | ||
Zhang B, Guo DD, Zheng JY, Wu YA, Ya W. Expression of KLF6-SV2 in colorectal cancer and its impact on proliferation and apoptosis. Eur J Cancer Prev. 2018;27(1):20–26. | ||
Hatami R, Sieuwerts AM, Izadmehr S, et al. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci Transl Med. 2013;5169(169):ra123. | ||
Teixeira MS, Camacho-Vanegas O, Fernandez Y, et al. KLF6 allelic loss is associated with tumor recurrence and markedly decreased survival in head and neck squamous cell carcinoma. Int J Cancer. 2007;121(9):1976–1983. | ||
Difeo A, Narla G, Hirshfeld J, et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006;12(12):3730–3739. | ||
Difeo A, Feld L, Rodriguez E, et al. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 2008;68(4):965–970. | ||
Sangodkar J, Difeo A, Feld L, et al. Targeted reduction of KLF6-SV1 restores chemotherapy sensitivity in resistant lung adenocarcinoma. Lung Cancer. 2009;662(3):292–297. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.