Back to Journals » OncoTargets and Therapy » Volume 12
KIF5B-RET fusion gene and its correlation with clinicopathological and prognosis features in lung cancer: a meta-analysis
Authors Cong XF, Yang L, Chen C, Liu Z
Received 4 September 2018
Accepted for publication 6 February 2019
Published 11 June 2019 Volume 2019:12 Pages 4533—4542
DOI https://doi.org/10.2147/OTT.S186361
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Xiao-Feng Cong, Lei Yang, Chen Chen, Ziling Liu
Department of Oncology, The First Hospital of Jilin University, Changchun, Jilin 130021, People’s Republic of China
Background: The KIF5B-RET fusion gene is a novel oncogene that has been observed in a subset of lung cancers in recent years. However, the results of related epidemiological studies remain unclear. Thus, a meta-analysis was conducted to evaluate the correlation of KIF5B-RET expression based on RT-PCR detection with clinicopathological features and prognosis of lung cancer.
Methods: The PubMed, Google Scholar, Wiley Online, SpringerLink and Chinese National Knowledge Infrastructure databases were searched to identify the eligible studies. The association of the occurrence of KIF5B-RET fusion gene in lung cancer with age, gender, smoking status, histology type, differentiation and TNM stage was analyzed. HR, overall survival (OS) and progression-free survival (PFS) were used to describe the prognosis of patients with lung cancer. The OR and 95% CI were calculated to assess the correlations. Random- and fixed-effects models were used to analyze the data.
Results: A total of 13 studies, which included 8,859 lung cancer patients, were included in the study based on the inclusion criteria. A total of 121 patients with positive KIF5B-RET fusion gene status were detected, with a positive expression rate of 1.36%. KIF5B-RET fusion gene status was identified at significantly higher frequencies in female (OR=0.67, 95% CI=0.48–0.94) than male patients, and the same trend was found in young (<60 years) patients (OR=0.08, 95% CI=0.01–0.45) compared with old patients (≥60 years). No differences were found in the TNM stage, histology, differentiation and smoking. Based on the prognosis, no difference was found between the status of the positive and negative KIF5B-RET fusion genes in OS and PFS of patients.
Conclusion: The KIF5B-RET fusion gene occurred predominantly in young female patients with lung cancer. However, the relationship between the expression of the fusion gene and the prognosis of lung patients remains unclear.
Keywords: pathological parameters, KIF5B-RET, lung cancer, fusion gene
Introduction
Research has shown that lung cancer is one of the most fatal tumors among the various malignant tumors, resulting in more than one million deaths every year worldwide.1,2 In China, lung cancer-related mortality ranks first among malignant tumors.3 The vast majority of lung cancer patients have deteriorated at the time of initial diagnosis, making it difficult for them to undergo surgery. In recent years, chemotherapy has remained the main treatment for lung cancer. Despite continuous improvements in treatment technology, the prognosis of lung cancer remains poor, and the 5-year survival rate is only approximately 15%.4,5
Smoking is a major cause of lung cancer. More than 161,000 lung cancer deaths are projected to occur in the USA in 2008. Of these, an estimated 10%–15% will be caused by factors other than active smoking.6 The majority of lung cancer patients without a history of smoking develop cancer because of cancer-related driver genes.7 In the past decade, with the development of tumor molecular biology, a series of driver genes related to non-small-cell lung cancer (NSCLC) has been discovered, and these genes include EGFR, KRAS and EML4-ALK, which are often referred to as lung cancer treatment targets.8 However, in more than 40% of NSCLC patients, the driver genes are not determined.
Since 2012, approximately 4.3%–8% of patients with lung adenocarcinoma have presented with RET gene rearrangement, whereas the RET gene encodes RET receptor tyrosine kinase.9 RET fusion genes, including kinesin family member 5B (KIF5B), coiled-coil domain-containing protein 6 (CDC6), nuclear receptor coactivator 4 (NCOA4) and tripartite motif containing 33 (TRIM33), have been discovered, and KIF5B-RET is one of the most important genotypes.10 The KIF5B-RET fusion gene was first discovered in the liver metastases of an NSCLC patient in 2011, and has been considered to be another important tyrosine kinase inhibitor (TKI) target for NSCLC.11 The RET fusion gene can induce the occurrence of thyroid cancer, which is controlled by tumor progression via the RET inhibitor, but it has been rarely observed in lung cancer.12,13 KIF5B-RET is not expressed in normal lung tissue but is highly expressed in lung cancer tissues.14 Patients with the KIF5B-RET genotype have unique clinical features, suggesting that the target may be a more specific molecular marker in NSCLC. The KIF5B-RET fusion gene is a chimeric tyrosine kinase, consisting of 638 N-terminal amino acid residues of the KIF5B protein and 402 C-terminal amino acid residues of the RET protein, including the RET gene coding.15,16 A structural region is present, having tyrosine protein kinase activity, a supercoiled domain and a motor domain encoded by the KIF5B gene.17 The supercoiled domain of the KIF5B-RET fusion protein can undergo homodimerization, activate the intracellular tyrosine kinase protein and open the oncogenic signaling pathway, and ultimately regulates cell growth and differentiation mainly through the Ras–Raf–MAPK and PI3K–Akt pathway.17 In summary, the related targeted drugs, diagnostic methods, clinical trials and transformational studies of KIF5B-RET/EML4-ALK fusion genes need further study.18
In this study, a meta-analysis was employed to study the expression of KIF5B-RET in lung cancer and related pathological data of lung cancer patients, including age, sex, TNM stage, smoking status, histological classification and differentiation. The expression of RET in patients was also studied. The prognosis of lung cancer patients was analyzed to predict the expression of the KIF5B-RET fusion gene for the treatment of lung cancer.
Methods
Data sources and search strategy
This meta-analysis was performed according to the PRISMA recommendations. The articles were retrieved by PubMed, Google Scholar, Wiley Online, SpringerLink and China National Knowledge Infrastructure and were collected from January 2012 to August 2018. The search keywords adoption strategy included (“Lung Cancer” or “Lung Neoplasms” or “Pulmonary Neoplasm”) and (“rearranged during transfection” or “RET”) and (“kinesin family member 5B” or “KIF5B” or “KIF5B-RET”) and/or “prognosis”. The search was restricted to RT-PCR detection studies, which were published in English and other languages. Detailed retrieval strategies are presented in the Supplementary material.
Inclusion and exclusion criteria
The inclusion criteria for the article included the following: 1) patients with pathology confirmed as lung cancer; 2) with available patient epidemiological and clinicopathological data or patient prognosis; 3) with available data forms for analysis and 4) articles published in English or Chinese. The exclusion criteria included the following: 1) patients with pathology confirmed as benign lung tumors; 2) no relevant pathology and patient prognosis data; 3) article data could not be used for statistical analysis; 4) non-human lung cancer tissues and 5) review studies and articles published with the same patient data. In cases of repeated publication of patient data, we used the latest article version for statistical analysis.
Data extraction
The data extracted from the article were collected independently by two investigators, and disputes were resolved through negotiation. Table 1 summarizes the basic data of each article, including the author’s first name, year of publication, area of patient data collection, detection method of related genes and positive expression rate of gene fusion.
  | Table 1 Basic information on the patients with lung cancer |
Statistical analyses
Stata 12.0 (StataCorp., College Station, TX, USA) was used to analyze the data related to lung cancer patients. Engauge Digitizer software was used to organize the survival curves of patient overall survival (OS) and progression-free survival (PFS). The summarized indicators included the patient’s KIF5B-RET expression rate, patient-related clinicopathological parameters and patient prognosis. The OR and corresponding 95% CI were used to describe the patient’s clinicopathological parameters. HRs, OS and PFS were used to analyze the prognosis of patients. The selection of the effect model of the data was based on the specific value of I2. When I2≤50%, the data have moderate or low heterogeneity, and the fixed-effects model is used for simulation. When I2>50%, the data have high heterogeneity, and the random-effects model is used for simulation. The publication bias for patient prognosis was simulated using a funnel plot and Egger’s test.
Results
Study selection and characteristics of included studies
A total of 1,028 articles were retrieved from relevant databases. Through the preliminary reading and analysis of the title and abstract, 887 articles were excluded because of a lack of significant relevance to the paper. The full text of the remaining 141 articles was downloaded and viewed, excluding the abstract, and the KIF5B-RET-related factors in animal lung cancer models and cell experiments were detected. Further analysis of the articles excluded 16 articles that could not be extracted and published using the same patient data. Finally, 13 related articles were included. The search results are presented in Figure 1.
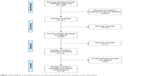  | Figure 1 Flow diagram of the study selection procedure and specific reasons for exclusion in the meta-analysis. |
A total of 8,859 patients were included in the 13 articles, including 169 patients with RET-positive disease, with a positive expression rate of 1.91%. A total of 121 patients had the KIF5B-RET fusion gene, with a positive expression rate of 1.36%.
Clinicopathological parameters
The KIF5B-RET fusion gene was identified at significantly lower frequencies in male (OR=0.67, 95% CI=0.48–0.94; Z=2.30, p=0.022) than in female patients, and the same trend was present in older (≥60 years) patients (OR=0.08, 95% CI=0.01–0.45; Z=2.87, p=0.004) compared with younger patients (<60 years) (all p<0.05) (Figures 2 and 3).
No differences were found in the TNM stage (I+II vs III+IV; Z=1.63, p=0.104; I2=0.0%, p=0.998), histology (adenocarcinoma of the lung vs non-adenocarcinoma; Z=1.41, p=0.158; I2=57.7%, p=0.021), differentiation (High vs Poor + Mod; Z=1.00, p=0.316; I2=89.7%, p=0.002) and smoking (Yes vs No; Z=1.88, p=0.061; I2=72.1%, p=0.000). The details are shown in Figures 2 and 3.
Prognosis
We analyzed six articles reporting OS and two articles reporting PFS for the KIF5B-RET fusion gene. The results showed no difference in OS and PFS between the positive and negative RET fusion genes (OS: Z=1.6, p=0.109; I2=67.3%, p=0.009; PFS: Z=1.75, p=0.080; I2=0.0%, p=0.723) (Figure 4).
Publication bias
Egger’s test showed that no publication bias was observed in the comparison between the positive and negative RET fusion gene. Funnel plots are presented in Figures 5 and 6. A graph of risk of bias (Figure 7) and a summary of risk of bias (Figure 8) were generated to describe the risk of bias of each study.
  | Figure 5 Egger’s publication bias plot of overall survival. |
  | Figure 6 Funnel plot of overall survival. |
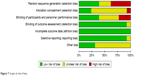  | Figure 7 Graph of risk of bias. |
  | Figure 8 Summary of risk of bias. |
Discussion
In recent years, genetic testing has been used to divide the same pathological type of lung cancer into various “molecular subtypes”, and individualized treatments can be achieved by selecting molecularly targeted drugs.19 The KIF5B-RET fusion gene is an independent and key molecular target for the development and progression of lung cancer, mainly in patients with EGFR, KRAS wild-type, non-smoker and young male lung adenocarcinoma.20–22 Several multi-target molecular targeted drugs are available internationally that may provide individualized treatment for patients with RET fusion genes.23 The emergence of the molecular subtype of KIF5B-RET fusion genotype lung cancer has further improved the pattern of NSCLC molecular typing diagnosis and treatment.
A total of 13 articles meeting the inclusion criteria were included in this meta-analysis, including 8,859 patients with lung cancer. Among them, 169 patients were RET positive, with a positive expression rate of 1.91%, whereas 121 patients had the KIF5B-RET fusion gene, with a positive expression rate of 1.36%. Analysis of clinicopathological parameters showed that the KIF5B-RET fusion gene was differentially expressed according to age and sex (p<0.05). In the TNM stage, histology, differentiation and smoking, RET was expressed between the higher frequencies and lower frequencies groups, and no difference was observed in the expression (all p-values >0.05). In particular, smoking was significantly associated with lung cancer. The KIF5B-RET fusion gene was originally found in the liver metastases of a lung cancer patient who did not have a history of smoking. Increasing evidence has indicated that the KIF5B-RET fusion gene is present in lung cancer patients who do not smoke or only smoke lightly. Kohno et al found that the proportion of non-smokers in patients with KIF5B-RET fusion gene-positive lung adenocarcinoma was 85.7% (6/7).11 Wang et al examined the status of the RET fusion gene in 936 patients with NSCLC who underwent lung resection in China.24 The proportion of non-smokers in lung adenocarcinoma patients with positive RET fusion gene was 81.8% (9/11). In KIF5B, the proportion of non-smokers in patients with positive RET fusion gene lung adenocarcinoma was 63.6% (7/11). These findings suggest that the KIF5B-RET fusion gene is more common in patients who do not smoke or only smoke lightly. Furthermore, the above data objectively support the research results of this paper.
Specific targeted drugs against the KIF5B-RET fusion gene have not been developed, but some TKIs that inhibit the activity of RET proteins have been widely clinically tested in thyroid cancer, and the US Food and Drug Administration has approved vandetanib for the treatment of hereditary thyroid gland medullary carcinoma. Kohno et al11 showed that vandetanib can inhibit the growth of NIH3T3 lung cancer cells containing the KIF5B-RET fusion gene. Lipson et al found that Ba/F3 cells transfected with the KIF5B-RET fusion gene showed high expression and phosphorylation activation of RET, whereas in vitro studies found that multi-targeted drugs, sunitinib, sorafenib and vandetanib, are effective in inhibiting the proliferation of this cell, whereas gefitinib did not have this effect.25
Lung cancer driver genes have been a hotspot in NSCLC-targeted therapy research. TKI treatment targeting the EGFR mutation and EML4-ALK fusion gene has introduced new ideas for NSCLC treatment.26 However, more than 40% of NSCLC-driven genes remain unclear.27 The discovery of the KIF5B-RET fusion gene has injected new vitality into the field of lung cancer research. However, to establish a diagnosis and treatment model that is truly similar to the current EGFR mutation, the ALK fusion gene requires a large amount of preclinical research and a high level of clinical evidence.
The results of this study showed that the expression of the KIF5B-RET fusion gene does not affect the patient’s OS and PFS. This finding may be partly due to the low expression rate of the KIF5B-RET fusion gene in lung cancer. Hence, more research data are needed to supplement this result. In the near future, we predict that researchers worldwide will continue to actively explore the diagnostic techniques and undertake clinical trials on KIF5B-RET fusion genes.
Disclosure
The authors report no conflicts of interest in this work.
References
Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011;32(4):605–644. doi:10.1016/j.ccm.2011.09.001 | ||
Miller RA, Cagle PT. Lung Cancer Epidemiology and Demographics. In: Cagle PT, Allen TC, Beasley MB, Chirieac LR, Dacic S, Borczuk AC, Kerr KM, Sholl LM, Portier B, Bernicker EH, editors. Precision Molecular Pathology of Lung Cancer. Houston: Molecular Pathology Library; 2018:15–17. | ||
Chen W, Zheng R, Zeng H, Zhang S. Epidemiology of lung cancer in China. Thoracic Cancer. 2015;6(2):209–215. doi:10.1111/1759-7714.12169 | ||
Shi L, Wang Z, Yu T, Xia L. Chronic disease and its therapeutic drugs may effect on the postoperative 5-year survival rate of lung cancer patients. J Chin Pharm Sci. 2017;26(2). doi:10.5246/jcps.2017.02.014 | ||
Noori FF, Karippot A, Kabak B, et al. 5-year survival rate of lung cancer patients in an inner city hospital. Chest. 2009;136(4):21. doi:10.1378/chest.136.4_MeetingAbstracts.21S-c | ||
Jonathan MS, Erika AT, Paolo B, et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15(18):5626–5645. doi:10.1158/1078-0432.CCR-09-0376 | ||
Aisner DL, Sholl LM, Berry L, et al; LCMC2 investigators. The impact of smoking and TP53 mutations in lung adenocarcinoma patients with targetable mutations – the Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res. 2017;24(5):1038–1047. | ||
Wang J, Suqin YI, Wang H, Zhou J, Guo F, Lab C. The analysis of c-Met, EGFR, K-Ras, and EML4-ALK in non-small cell lung cancer. Chin Clin Oncol. 2015;20(10):902–908. | ||
Cai W, Su C, Li X, et al. KIF5B-RET fusions in Chinese patients with non-small cell lung cancer. Cancer. 2013;119(8):1486–1494. doi:10.1002/cncr.27940 | ||
Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non–small cell lung cancer. Clin Cancer Res. 2008;14(10):2895–2899. doi:10.1158/1078-0432.CCR-07-2248 | ||
Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18(3):375–377. doi:10.1038/nm.2644 | ||
Hart CD, De Boer RH. Profile of cabozantinib and its potential in the treatment of advanced medullary thyroid cancer. Onco Targets Ther. 2013;6(1):1–7. | ||
Samadi AK, Mukerji R, Shah A, Timmermann BN, Cohen MS. A novel RET inhibitor with potent efficacy against medullary thyroid cancer in vivo. Surgery. 2010;148(6):1228–1236. doi:10.1016/j.surg.2010.09.026 | ||
Andrew IS, Michael PC, Keith AC, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99(7):4465–4470. doi:10.1073/pnas.012025199 | ||
Sasaki H, Shimizu S, Tani Y, et al. RET expression and detection of KIF5B/RET gene rearrangements in Japanese lung cancer. Cancer Med. 2012;1(1):68–75. doi:10.1002/cam4.13 | ||
Huang Q, Schneeberger VE, Luetteke N, et al. Preclinical modeling of KIF5B-RET fusion lung adenocarcinoma. Mol Cancer Ther. 2016;15(10):2521. doi:10.1158/1535-7163.MCT-15-0003 | ||
Roskoski JR, Sadeghinejad A. Role of RET protein-tyrosine kinase inhibitors in the treatment RET-driven thyroid and lung cancers. Pharmacol Res. 2017;128:1. doi:10.1016/j.phrs.2017.12.021 | ||
Suehara Y, Arcila M, Wang L, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18(24):6599–6608. doi:10.1158/1078-0432.CCR-12-0838 | ||
Ni J, Li Z. Evaluation of three small molecular drugs for targeted therapy to treat nonsmall cell lung cancer. Chin Med J (Engl). 2016;129(3):332–340. doi:10.4103/0366-6999.174484 | ||
Borrelli N, Giannini R, Proietti A, et al. KIF5B/RET fusion gene analysis in a selected series of cytological specimens of EGFR, KRAS and EML4-ALK wild-type adenocarcinomas of the lung. Lung Cancer. 2013;81(3):377–381. doi:10.1016/j.lungcan.2013.06.026 | ||
Kim JO, Lee J, Shin JY, et al. KIF5B-RET fusion gene may coincide oncogenic mutations of EGFR or KRAS gene in lung adenocarcinomas. Diagn Pathol. 2015;10(1):143. doi:10.1186/s13000-015-0368-z | ||
Akcay S, Er Dedekarginoglu B. Smoking cessation in lung cancer. J Lung Cancer Diagn Treat. 2016;1:105. doi: 10.4172/jlcdt.1000105 | ||
Yano S, Takeuchi S, Nakagawa T, Yamada T. Ligand-triggered resistance to molecular targeted drugs in lung cancer: roles of hepatocyte growth factor and epidermal growth factor receptor ligands. Cancer Sci. 2012;103(7):1189–1194. doi:10.1111/j.1349-7006.2012.02279.x | ||
Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non–small-cell lung cancer. J Clin Oncol. 2012;30(35):4352–4359. doi:10.1200/JCO.2012.44.1477 | ||
Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18(3):382–384. | ||
Antonicelli A, Cafarotti S, Indini A, et al. EGFR-targeted therapy for non-small cell lung cancer: focus on EGFR oncogenic mutation. Int J Med Sci. 2013;10(3):320–330. doi:10.7150/ijms.4609 | ||
Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi:10.1016/S1470-2045(10)70087-5 | ||
Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18(3):378–381. doi:10.1038/nm.2658 | ||
Yokota K, Sasaki H, Okuda K, et al. KIF5B/RET fusion gene in surgically-treated adenocarcinoma of the lung. Oncol Rep. 2012;28(4):1187–1192. doi:10.3892/or.2012.1908 | ||
Yoo SS, Jin G, Jung HJ, et al. RET fusion genes in Korean non-small cell lung cancer. J Korean Med Sci. 2013;28(10):1555–1558. doi:10.3346/jkms.2013.28.10.1555 | ||
Tsuta K, Kohno T, Yoshida A, et al. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br J Cancer. 2014;110(6):1571–1578. doi:10.1038/bjc.2014.36 | ||
Pan Q, Wang Y, Chen J, et al. Investigation of the epidermal growth factor receptor mutation rate in non-small cell lung cancer patients and the analysis of associated risk factors using logistic regression. Oncol Lett. 2014;8(2):813–818. doi:10.3892/ol.2014.2160 | ||
Tsai TH, Wu SG, Hsieh MS, Yu CJ, Yang JC, Shih JY. Clinical and prognostic implications of RET rearrangements in metastatic lung adenocarcinoma patients with malignant pleural effusion. Lung Cancer. 2015;88(2):208–214. doi:10.1016/j.lungcan.2015.02.018 | ||
Song Z, Yu X, Zhang Y. Clinicopathologic characteristics, genetic variability and therapeutic options of RET rearrangements patients in lung adenocarcinoma. Lung Cancer. 2016;101:16–21. doi:10.1016/j.lungcan.2016.09.002 | ||
Yu T, Xue S, Jia C, Wang R. KIF5B-RET and EML4-ALK fusion gene expression status and survival analysis of stage IV Uygur NSCLC patients. J Pract Oncol. 2018. |
Supplementary material
Electronic search strategy
PICOS
P (patient or population): Lung Cancer
[MESH] (((((Lung Cancer) OR Pulmonary Neoplasm) OR Neoplasms, Lung) OR Neoplasms, Pulmonary) OR Cancer, Lung) OR Pulmonary Neoplasm.
I (intervention/exposure): RT-PCR detection the KIF5B-RET fusion gene expression
[MESH] ((((kinesin family member 5B) OR KIF5B protein, human) OR kinesin family member 5B, human) OR KIF5B-RET) OR KIF5B-RET fusion protein, human.
RET [MESH] (((((((Proto Oncogene Proteins) OR Proto-Oncogene Products, Cellular) OR Cellular Proto-Oncogene Products) OR RET) OR Proto Oncogene Products, Cellular) OR c-onc Proteins) OR Cellular Proto-Oncogene Proteins) OR Proto-Oncogene Proteins, Cellular.
C (comparison/control): Negative expression of KIF5B-RET fusion gene.
O (outcome): Clinicopathological (age, sex, TNM stage, smoking status, histological classification and differentiation); Prognosis (PFS, OS).
Prognosis [MESH] ((((Prognoses) OR Prognostic Factors) OR Factor, Prognostic) OR Prognostic Factor) OR Factors, Prognostic.
S (study design): Diagnostic study.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



