Back to Journals » International Journal of General Medicine » Volume 14
KDELC1 and TRMT1 Serve as Prognosis-Related SARS-CoV-2 Proteins Binding Human mRNAs and Promising Biomarkers in Clear Cell Renal Cell Carcinoma
Authors Li C, Yao Y, Long D, Lin X
Received 26 March 2021
Accepted for publication 17 May 2021
Published 14 June 2021 Volume 2021:14 Pages 2475—2490
DOI https://doi.org/10.2147/IJGM.S312416
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Canxuan Li,1,* Yuzhi Yao,2,3,* Dan Long,4 Xiaobin Lin2,5
1Department of Urology, The First Affiliated Hospital of Jinan University, Guangzhou, Guangdong, People’s Republic of China; 2Department of Breast Surgery and General Surgery, Guangzhou Women and Children’s Medical Center, Guangzhou, Guangdong, People’s Republic of China; 3Guangzhou Women and Children’s Medical Center, Department of Paediatric Surgery Clinic, Guangzhou, Guangdong, People’s Republic of China; 4Respiratory medicine, Shenshan Central Hospital, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Shanwei, Guangdong, People’s Republic of China; 5Department of Breast Surgery, The First Affiliated Hospital of Jinan University, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaobin Lin
Department of Breast Surgery and General Surgery, Guangzhou Women and Children’s Medical Center, Guangzhou, Guangdong, People’s Republic of China
Email [email protected]
Background: SARS-CoV-2 proteins binding human mRNAs (SPBRs) have been proven to regulate a variety of tumor-related functions in different types of cancer. However, their biological roles and potential mechanisms in clear cell renal cell carcinoma (ccRCC) are still elusive. Herein, we investigate the expression and prognostic value of SPBRs in ccRCC through bioinformatics methods.
Methods: Data downloaded from the Cancer Genome Atlas (TCGA) database was used to screen differentially expressed SPBRs (DE-SPBRs) between ccRCC samples and noncancerous samples. Metascape was utilized to perform function and pathway enrichment analyses of these DE-SPBRs. Kaplan–Meier method of overall survival (OS) was used to assess the prognostic value of DE-SPBRs in ccRCC patients. Univariate and multivariate Cox regression analyses were applied to identify candidate SPBRs, which were independently associated with overall survival of ccRCC patients. Subsequently, several internationally renowned databases were employed to conduct a comprehensive analysis of candidate SPBRs to further investigate their roles and mechanisms in ccRCC.
Results: A total of 33 DE-SPBRs, including 18 upregulated SPBRs and 17 downregulated SPBRs, were screened between ccRCC samples and noncancerous samples. Among them, two candidate SPBRs, KDELC1 and TRMT1, were identified. Additionally, we observed that upregulated KDELC1/TRMT1 expression in ccRCC at both gene and protein levels was significantly associated with clinicopathological features. Furthermore, we found that KDELC1/TRMT1 genetic mutation has an unfavorable influence on prognosis of patients with ccRCC. Functional enrichment analysis revealed that KDELC1/TRMT1 was closely enriched in several vital biological processes and pathways. Finally, we noticed that KDELC1/TRMT1 was remarkably associated with immune infiltrates.
Conclusion: In summary, we screened DE-SPBRs of ccRCC, which were enriched mainly in various biological and signaling pathways with tumor progression. Furthermore, we identified two candidate DE-SPBRs (KDELC1 and TRMT1), which could serve as promising biomarkers and therapeutic targets of patients with ccRCC.
Keywords: clear cell renal cell carcinoma, bioinformatics, SARS-CoV-2 proteins binding human mRNAs
Introduction
Clear cell renal cell carcinoma (ccRCC), originates from renal tubular epithelial cells, which is the most common histologic subtype of renal cell carcinoma (RCC) with an incidence accounting for about 75% of all cases.1 Surgical intervention is an effective treatment for ccRCC patients with early stage. However, owing to lack of specific signs and symptoms, majority of patients reach advanced stage of the cancer at the first diagnosis with unresectable primary tumors.2 Although significant progress has been made in diagnostic techniques and therapies, most patients’ prognosis remains poor, mainly due to the distant metastases and recurrence.3,4 To date, causing by the high recurrence and incidence in the ccRCC, it is quite urgent to find promising biomarkers and therapeutic targets for ccRCC patients. In recent years, targeted therapy has been proposed as a promising anticancer strategy. Many molecular studies have focused on early diagnosis and prognosis with iconic targets, but the missed diagnosis rates still very high.5 Therefore, searching out novel therapeutic biomarkers associated with ccRCC is crucial for targeted therapy. SARS-CoV-2 is the culprit of the coronavirus disease 2019 (COVID-19), which has led to global health emergencies and social and economic disruptions worldwide.6,7 Studies have shown that regardless of receiving any special treatment for COVID-19, cancer patients seem to show an increased risk of death from COVID-19.8 Angiotensin-converting enzyme 2 (ACE2) has shown strong binding to SARS-CoV-2 and has been confirmed as a functional host cell receptor.9 ACE2 is a member of the renin-angiotensin system and usually expressed in many organs and its overexpression seemed to be more susceptible to SARS-CoV-2 infection.10 Although there is no direct evidence of an association between clear cell renal cell carcinoma and SARS-CoV-2, renal tubular epithelial cells express more ACE2,11 suggesting that they are susceptible to SARS-CoV-2. ACEis can potentially inhibit cancer growth, reduce the potential for metastasis, and improve survival in patients with RCC,12 while increased ACE activity is associated with higher tumor grade.13 Mechanically, the therapeutic use of ACEis in RCC is of great significance for the treatment of RCC patients infected with COVID-19.14 Because of the increase in ACE2, the increase in ACE2 to which the virus can bind has the potential to increase the susceptibility of RCC patients to more severe diseases.
To date, at least 330 high-confidence SARS-CoV-2 proteins binding human mRNAs (SPBRs) have been identified in the human genome. They could combine with their target RNAs to regulate many physiological and biochemical processes, such as coronavirus replication. lipoprotein metabolism, DNA replication, RNA processing and regulation, and epigenetic regulation of gene expression.15,16 These abilities implicated that the abnormal expression of SPBRs is related with the initiation and progression of various human cancers. For instance, TRIM59 by upregulating cell cycle–related proteins to facilitates the proliferation and migration of Non-Small Cell Lung Cancer Cells.17 In glioblastoma, LncRNA PLAC2 blocks cell cycle progression by down-regulating RPL36 expression.18 LOX was highly expressed in colorectal cancer samples than in adjacent normal samples, and upregulated LOX indicated advanced clinical stages, deeper tumor invasion, and more metastatic lymph nodes.19 Despite the fact that SPBRs closely participated in the occurrence and progression of different types of human cancers, little is known about the function of most SARS-CoV-2 proteins binding human mRNAs (SPBRs) in ccRCC. An integrated study will help us understand the specific functional roles of SPBRs in ccRCC.
Gene expression data and corresponding clinicopathological information of ccRCC, which was obtained from The Cancer Genome Atlas (TCGA) database, was used to screened prognosis related differentially expressed SPBRs among cancerous tissues and healthy renal controls via comprehensive bioinformatics methods. Among them, two independent prognosis factors (KDELC1 and TRMT1) were identified using Cox analysis. Then, several universally acknowledged databases were employed to explore mRNA and protein expression levels, clinicopathological features correlation, genetic variations, gene–gene interaction network, functional enrichment and TIICs analyses of KDELC1 and TRMT1 in ccRCC. Taken together, our results demonstrated that KDELC1 and TRMT1 were associated with the occurrence and progression of ccRCC and could be recognized as promising biomarkers for ccRCC, and targeting SARS-CoV-2 proteins binding human mRNAs may be a new anti-tumor treatment strategy of ccRCC and will provide a good reference between ccRCC research and SARS-CoV-2.
Materials and Methods
Data Collection and Identification of Differentially Expressed SPBRs
TCGA (https://portal.gdc.cancer.gov/)20 database is an comprehensive database platform and the data that it contains is publicly available. In this work, we downloaded the RNA sequencing dataset of 72 normal renal samples and 539 KIRC samples. Duplicate samples and any samples with unavailable or unknown survival time data will be excluded, and finally we obtained RNA sequencing profile of 72 noncancerous specimens and 530 ccRCC specimens. Furthermore, clinical and pathological characteristics of patients with ccRCC, including age, gender, histological grade, clinical stage, and TMN stage, were recorded in Table 1. Subsequently, differential SPBRs expression between ccRCC samples and non-cancerous renal samples was screened using the edgeR package (www.bioconductor.org/packages/release/bioc/html/edgeR.html).21 A total of 334 SPBRs were included in the work based on the results of the literature review (Supplementary_Table S1). SPBRs that met the specific cut-off criteria of adjusted P < 0.05 and |log2fold change (FC)|≥1 were considered as differentially expressed SPBRs (DE-SPBRs).
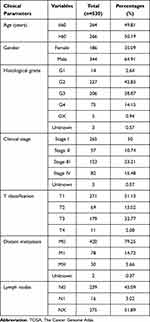 |
Table 1 Characteristics of Clear Cell Renal Cell Carcinoma Patients in the TCGA Database |
Functional Enrichment Analysis of DE-SPBRs
Metascape (http://metascape.org) is a web-based portal designed to perform functional enrichment, interactome analysis, gene annotation, and membership search.22 In this study, Metascape was used to analyze Function and pathway enrichment analyses of DE-SPBRs, gene terms with P-value <0.01, minimum count of 3, and enrichment factor of >1.5 were regarded significantly.
Identification of Prognosis-Related DE-SPBRs
To evaluate the prognostic value of these DE-SPBRs, overall survival analysis of each DE-SPBRs was performed through the survival package of R software. All patients were divided into high- and low-expression subgroups according to the median expression value. DE-SPBRs that met the cut-off criteria of P < 0.05 were considered as prognosis-related DE-SPBRs. Finally, univariate and multivariate cox analyses were performed to determine independent prognostic predictors with the overall survival of patients with ccRCC. These independent prognostic DE-SPBRs were considered as candidate DE-SPBRs.
Expression Validation and Clinicopathological Analysis of Candidate DE-SPBRs in ccRCC
Chip data derived from ccRCC patients, downloaded from GSE15641, GSE36895, GSE53757, and GSE66272, were used for expression validation. The latter three datasets were from the same platform: GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array; contained 29 tumor samples and 23 normal samples,23 72 tumor samples and 72 normal tissues,24 and 27 tumor samples and 27 normal tissues,25,26 respectively. GSE15641 dataset was from the Platform: GPL96 [HG-U133A] Affymetrix Human Genome U133A Array, which contained 32 tumor samples and 23 adjacent non-tumor renal tissues.27 Furthermore, the protein expression for colon cancer, breast cancer, ovarian cancer, clear cell renal cell carcinoma, and uterine corpus endometrial carcinoma can be obtained from the “CPTAC analysis” module of UALCAN (http://ualcan.path.uab.edu).28 In this study, candidate DE-SPBRs protein expression pattern in ccRCC and noncancerous tissues was obtained from the UALCAN web tool. Additionally, we selected representative protein images to analyze the differences in staining at the protein level of candidate genes from the HPA database.29 Additionally, the Wilcoxon signed-rank test was utilized to explore the association between clinicopathological factors and candidate DE-SPBRs.
cBioPortal for Cancer Genomics Database
The cBioPortal for cancer genomics (http://www.cbioportal.org/) is a powerful open-source database, which can provide multidimensional cancer genomics and clinical data retrieved from the TCGA database. Here, the kidney renal clear cell carcinoma (KIRC) dataset, including data from 537 cases with pathology reports, was selected for further analyses of candidate DE-SPBRs using cBioPortal database. The cBioPortal Cancer Genomics resource was used to assess the alteration frequency of the candidate DE-SPBRs and its influence on prognosis of ccRCC cases.
GENEMANIA Database
The GeneMANIA is a prediction server, which could find other genes that are related to a set of input genes, using a very large set of functional association data. Association data include protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity. In the work, we use GeneMANIA to construct a gene–gene network. Furthermore, to understand the potential mechanisms of candidate DE-SPBRs in ccRCC, we carried out function and pathway enrichment analyses to explore the potential biological mechanisms, gene terms with P-value <0.01, minimum count of 3, and enrichment factor of >1.5 were regarded significantly.
TIMER Database
The Tumor Immunity Resource (TIMER) (https://cistrome.shinyapps.io/timer/) database contains mRNA sequencing data for 10,897 samples of 32 human cancers from the Cancer Genome Atlas (TCGA). It is an ideal tool for systematic analysis of immune infiltration in various cancer types. In this study, we assessed the candidate DE-SPBRs expression with the abundance of different immune infiltrating cells.
Statistical Analysis
The Wilcoxon signed-rank test was utilized to compare the expression levels of the candidate DE-SPBRs in ccRCC samples with that in normal renal samples, and to explore the association between clinicopathological variables and the candidate DE-SPBRs. Kaplan–Meier survival method with Log rank test was used to estimate the effect of the candidate DE-SPBRs on overall survival of ccRCC patients. Univariate Cox regression and multivariate Cox analyses were performed to screed the independent prognostic factors. P < 0.05 was regarded as statistically significant. All statistical analyses were carried out using R 3.6.1 software.
Results
Identification of DE-SPBRs
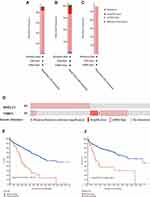
The edgeR R package was employed to screen the DE-SPBRs between ccRCC tissues and noncancerous tissues. A total of 334 SPBRs were included in this work, and 35 DE-SPBRs, including 18 upregulated SPBRs and 17 downregulated RBPs, were identified between ccRCC samples and noncancerous samples with the setting standard |log2FC| ≥1 and the adjusted P-value < 0.05 (Table 2). Additionally, Figure 1A and B shows the heat map and volcano map of DE-SPBRs, respectively.
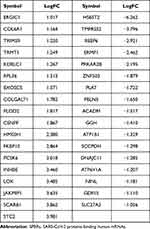 |
Table 2 Identification of Differentially Expressed SPBRs |
 |
Figure 1 Identification of differentially expressed SARS-CoV-2 proteins binding human mRNAs (DE-SPBRs). (A) Heat map of DE-SPBRs; (B) volcano plot of DE-SPBRs. |
Function and Pathway Enrichment Analyses of DE-SPBRs
To investigate the biological mechanisms of DE-SPBRs in ccRCC, we performed function and pathway enrichment analyses of DE-SPBRs using the Metascape database.
As can be seen from Figure 2, gene ontology (GO) indicated that DE-SPBRs were mainly involved in collagen formation, response to oxygen levels, NABA ECM regulators, response to peptide hormone, transport of small molecules, glycoprotein metabolic process, G2/M transition, cellular modified amino acid metabolic process, cellular lipid catabolic process, hormone metabolic process and signaling by receptor tyrosine kinases.
Identification of Prognosis-Related DE-SPBRs
To investigate the prognostic value of DE-SPBRs in ccRCC, Kaplan–Meier curve for overall survival of ccRCC patients was calculated according to the low and high expressions of each DE-SPBRs. Twenty DE-SPBRs showed statistically favorable/unfavorable effect on prognosis of patients with ccRCC. As shown in Figure 3, Kaplan–Meier curves showed that CENPF, COL6A1, COLGALT1, EXOSC5, FKBP10, INHBE, KDELC1, PLOD2, RPL36, TRIM59, TRMT1, PPKAR2B and REEP6 have lower percent overall survival in the high-expression group compared to the low-expression group (Figure 3A–I, 3K–L, and 3R–S). Compared with the high-expression group, SCARB1, ACADM, ATP1B1, ATP6V1A, ERMP1, GGH, and SLC27A2 have an unfavorable overall survival in low-expression group (Figure 3J, M–Q, and T). Moreover, as shown in Table 3, in Univariate analysis, ERMP1 expression (HR (Hazard Ratio) = 0.681, p < 0.05), REEP6 expression (HR = 1.162, p < 0.05), COLGALT1 expression (HR = 3.011, p < 0.001), ACADM expression (HR = 0.470, p < 0.001), ATP1B1 expression (HR = 0.615, p < 0.001), FKBP10 expression (HR = 1.474, p < 0.001), TRIM59 expression (HR = 1.705, p < 0.01), EXOSC5 expression (HR = 1.371, p < 0.05), GGH expression (HR = 0.731, p = p < 0.01), TRMT1 expression (HR = 2.032, p < 0.001), KDELC1 expression (HR = 1.536, p < 0.01), PLOD2 expression (HR = 1.234, p < 0.05), CENPF expression (HR = 1.640, p < 0.001), ATP6V1A expression (HR = 0.686, p < 0.05), RPL36 expression (HR = 1.293, p < 0.05), INHBE expression (HR = 1.198, p < 0.001), COL6A1 expression (HR = 1.892, p < 0.001), SLC27A2 expression (HR = 0.749, p < 0.001), age (HR = 1.023; p < 0.05), histological grade (HR = 2.242; p < 0.001), clinical stage (HR = 1.862; p < 0.001), T classification (HR=1.943; p < 0.001), Lymph nodes (HR=2.932; p < 0.01) and distant metastasis (HR = 4.073; p < 0.001) were associated with poorer overall survival of patients with ccRCC. Furthermore, multivariate survival model after variable selection indicated that age (HR = 1.052; p < 0.001), distant metastasis (HR = 3.411; p < 0.05), KDELC1 expression (HR = 2.003, p < 0.001) and TRMT1 expression (HR = 2.423, p < 0.01) of patients with ccRCC were independently associated with unfavorable OS (Figure 4 and Table 3). The above results showed that KDELC1 and TRMT1 could serve as individual predictors for poor prognosis in ccRCC.
 |
Table 3 Univariate Cox Regression and Multivariate Cox Regression Analyses for Identification of Candidate SARS-CoV-2 Proteins Binding Human mRNAs |
 |
Figure 4 Multivariate Cox regression analysis to identify prognosis-related SARS-CoV-2 proteins binding human mRNAs (SPBRs). ***, P < 0.001. |
KDELC1 and TRMT1 Were Highly Expressed in ccRCC
As shown in Figure 5A–H, KDELC1 and TRMT1 overexpression were observed in ccRCC, according to GSE15641, GSE36895, GSE53757 and GSE66272 databases (P<0.001). Additionally, to determine the proteomic expression of KDELC1 and TRMT1 in ccRCC, we used protein expression data from the CPTAC dataset to show that KDELC1 and TRMT1 were significantly upregulated in ccRCC tissues by comparison with normal renal tissues (p < 0.001) (Figure 5I–J). Representative immunohistochemical staining showed that tumor tissue had much higher expression of KDELC1 than normal renal tissue (Figure 5K). However, no immunohistochemical staining difference was found between tumor and normal tissues for TRMT1 (Figure 5L).
The Expression of KDELC1/TRMT1 and Its Relationship with Clinicopathological Features
We evaluated the association between KDELC1/TRMT1 expression levels and different clinicopathological variables of ccRCC patients. The results are described in boxplot Figure 6, we found that overexpression of KDELC1 was significantly associated with histological grade (p < 0.01), clinical stage (p < 0.01), T classification (p < 0.05), and distant metastasis (p < 0.001). In addition, upregulated expression of TRMT1 was significantly clinical stage (p < 0.05), T classification (p < 0.05), and distant metastasis (p < 0.05). The results showed that increased KDELC1/TRMT1 expression indicates a worse clinical overcome.
KDELC1 and TRMT1 Genetic Alteration Analysis
Alteration frequency of KDELC1 and TRMT1 mutations in ccRCC was analyzed using cBioPortal database. A total of 537 cases from three datasets of KIRC were analyzed. Overview of the analyses of genetic variations in KDELC1 and TRMT1 was shown in Figure 7A. KDELC1 and TRMT1 were altered in 49/537 (9.12%) of ccRCC patient samples. These alterations were mRNA high in 44 cases (8.19%), amplification in 3 cases (0.56%), mutation in 1 case (0.19%) and multiple alterations in 1 case (0.19%). Furthermore, the analysis of genetic variations in KDELC1 and TRMT1 was shown in Figure 7B and C; genetic alterations were 4.1% and 5.59%, respectively. Figure 7D shows the OncoPrint visual summary of alteration on a query of KDELC1 and TRMT1. Furthermore, Kaplan–Meier curves revealed that the cases with alterations in one of the query genes have worse overall survival and disease-free survival than those without alterations in any query genes (Both p-values <0.001) (Figure 7E–F). The results indicated that KDELC1 and TRMT1 genetic alteration could have an unfavorable effect on prognosis of patients with ccRCC.
Co-Expression and Functional Enrichment Analyses
Genes significantly associated with KDELC1 and TRMT1 were analyzed using the GeneMANIA database (Figure 8A). To explore the potential mechanism of KDELC1 and TRMT1, we used the Metascape database to conduct function enrichment analysis. The results indicated that KDELC1/TRMT1 and their functional partners mainly enriched in several vital biological processes and pathways, such as ncRNA processing, RNA modification, protein O-linked glycosylation via serine, regulation of RNA splicing and regulation of reproductive process (Figure 8B–D).
TIMER Analysis
Accumulating evidence indicated that tumor-infiltrating lymphocytes could utilize to predict sentinel lymph node status and survival in tumors. Hence, we further analyzed the correlation between KDELC1 and TRMT1 expression with tumor-infiltrating immune cells. As illustrated in Figure 9, elevated KDELC1 was negatively correlated with tumor purity (r=−0.11, p < 0.001) in ccRCC and significantly associated with infiltrating levels of B cells (r=0.154, p < 0.001), CD4+ T (r=0.192, p < 0.001), macrophages (r=0.319, p < 0.001), neutrophils (r=0.343, p < 0.001), and dendritic cells (r=0.215, p < 0.001). However, increased TRMT1 was significantly associated with infiltrating levels of B cells (r=−0.253, p < 0.001), CD8+ T cells (r=−0.117, p < 0.05), CD4+ T (r=0.22, p < 0.001), macrophages (r=−0.21, p < 0.001), and dendritic cells (r=−0.182, p < 0.001). These findings suggested that KDELC1 and TRMT1 may play a crucial role in the ccRCC microenvironment.
 |
Figure 9 The correlation between KDELC1/TRMT1 expression with immune infiltration levels and in clear cell renal cell carcinoma. (A) KDELC1; (B) TRMT1. |
Discussion
Herein, we screened 35 DE-SPBRs between ccRCC samples and heathy normal controls from the TCGA database. Based on function and pathway enrichment analyses, we found that these DE-SPBRs involved in collagen formation, ECM regulators, G2/M transition, collagen is the major component of the cancer microenvironment and is involved in tumor fibrosis, tumor cells could regulate collagen biosynthesis through mutated genes, transcription factors, signaling pathways and receptors; additionally, collagen also can affect the behavior of tumor cells through integral proteins, discoidal protein structural domain receptors, tyrosine kinase receptors and several signaling pathways.30 The composition and organization of the ECM is dynamically regulated in both time and space to control cell behavior and differentiation, but imbalances in the dynamics of the ECM contribute to the progression of diseases such as cancer.31 Dysregulation of the cell cycle is one of the reasons why cancer cells divide abnormally, and the acceleration and activation of G2/M transition helps to enhance cancer cell proliferation.32 Furthermore, we found that these DE-SPBRs also participated in the metabolism of certain substances, including glycoprotein, cellular modified amino acid cellular lipid and hormone metabolic processes, suggesting that the metabolism of these substances may be involved in the development of ccRCC.
Subsequently, based on the Kaplan–Meier and cox regression analyses, two SPBRs (KDELC1 and TRMT1) independently associated with overall survival were selected for further comprehensive analysis. In this work, we test and verify the KDELC1 and TRMT1 mRNA expression in ccRCC tissue and its relationship with patient clinical outcomes. Results showed that KDELC1 and TRMT1 mRNA were significantly upregulated in cancer cases relative to normal heathy controls both in mRNA and protein levels and its high transcription levels were significantly associated with poor clinical outcomes. Furthermore, we noticed that mRNA high is the most frequent type of genetic alterations and significantly associated with a poor prognosis of patients with ccRCC, also indicating that genetic alterations might be responsible for the up-regulation of KDELC1 and TRMT1 gene. All above results suggested that KDELC1 and TRMT1 might play a vital role as a tumor-specific promoter in ccRCC.
KDELC1, also known as POGLUT2, encodes a protein product localized to the lumen of the endoplasmic reticulum.33 Although the direct role of KDELC1 in cancer has not been extensively studied, it has been shown to play an important role in the glycosylation of the heterodimeric Notch receptor, and the letter is significantly decisive in the initiation of Notch signaling that has great relevance to multiple aspects of cancer biology, including angiogenesis, tumor immunity and the maintenance of cancer stem-like cells.34 Additionally, as a novel glycosyltransferase, its abnormal dysregulation may be associated with the development of cancer.35 We thus speculate that it can potentially serve as a novel biomarker for ccRCC progression. TRMT1 is a tRNA modifying gene that acts as a dimethyltransferase, modifying a single guanine residue at position 26 of the tRNA. Notably, several studies demonstrated that tRNA modification is emerging as a novel mechanism of oncogenic regulation.36 Deve et al found that cells lacking TRMT1 show reduced proliferation rates, alterations in protein synthesis, and tolerance to oxidative stress.37 Additionally, Wang et al identified TRMT1 and other 17 genes that could predict lymph node metastasis of bladder cancer patients,38 suggesting that TRMT1 might play a vital role in carcinogenesis.
The relationship of the expression genes correlated with differentially expressed KDELC1 and TRMT1 was performed using the GENEMANIA database. The results showed that POGLUT1, ANKRD28, RRP9, TSR2, TRMT1L, METTL16, CTU1, TARBP2, RTFDC1, ERN1, KDELC2, ZNF598, THUMPD2, C1QBP, PDE3A, PDCD11, TPRKB, EXOSC10, CAPN15, and NUDC had different types of association with KDELC1/TRMT1, including physical Interactions, co-expression, predicted, co-localization, pathway, genetic interactions, shared protein domains. Then, we carried out functional enrichment analyses to explore the potential mechanism of KDELC1/TRMT1 and its functional partners. Mechanically, KDELC1 and TRMT1 affect the progression of ccRCC through several post-transcriptional events, such as ncRNA processing, RNA modification, protein O-linked glycosylation via serine, regulation of RNA splicing and regulation of reproductive process. Abnormal post-transcriptional events were the cause of the occurrence and development of cancer,39 which might lead to the activation of oncogenes or the inhibition of tumor suppressor genes. Abnormal post-transcriptional events might be the mechanism for KDELC1 and TRMT1 to promote ccRCC progression.
Immune cells in the tumor microenvironment regulate cancer progression and are attractive therapeutic targets.40,41 Therefore, for the first time, we explored the correlation between KDELC1/TRMT1 expression and tumor-infiltrating immune cells in ccRCC, and we found that KDELC1 was positively associated with infiltrating levels of B cells, CD4+ T, macrophages, neutrophils, and dendritic cells. However, TRMT1 was negatively related with infiltrating levels of B cells, CD8+ T cells macrophages, and dendritic cells, whereas positively associated with infiltrating levels of CD4+ T cells.
These findings suggested that KDELC1 and TRMT1 might play different and specific roles in the ccRCC microenvironment.
In conclusion, our study demonstrated that SARS-CoV-2 proteins binding human mRNAs might involve in the progression and prognosis of clear cell renal cell carcinoma. Among them, KDELC1 and TRMT1 are expected to become promising therapeutic targets in ccRCC treatment. To our best knowledge, this is the first work to investigate the roles of SPBRs in ccRCC. However, several limitations should not be ignored. First, this is a retrospective research so that selection bias is inevitable Second.
Secondly, certain clinical factors that can also influence prognosis, such as specific details of patient treatment, are missing from public databases. Finally, we cannot precisely describe the possible mechanisms of KDELC1 and TRMT1 in ccRCC. Therefore, further substantive research, such as cell-based protein expression assays, are required to confirm our conclusions.
Data Sharing Statement
The RNA-seq and clinical data of the 530 ccRCC patients from TCGA database were downloaded from https://portal.gdc.cancer.gov/.
Ethical Approval and Consent to Participate
The public database mentioned in this study is publicly available for re-analyzing, and no ethical approval was required by the local ethics committees, so that this study does not require ethics approval.
Acknowledgments
The authors thank Bo Zhao (Official Wechat Account: SCIPhD) of ShengXinZhuShou for suggestions on the manuscript.
Funding
This project is supported by the doctoral start-up fund of Guangzhou Women and Children’s Medical Center. Fund NO. 1600042-04.
Disclosure
The authors declare that they have no competing interests, and all authors confirm its accuracy.
References
1. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3(1):17009. doi:10.1038/nrdp.2017.9
2. Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67:507–524. doi:10.3322/caac.21411
3. Vera-Badillo FE, Templeton AJ, Duran I, et al. Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis. Eur Urol. 2015;67:740–749. doi:10.1016/j.eururo.2014.05.010
4. Fernández-Pello S, Hofmann F, Tahbaz R, et al. A systematic review and meta-analysis comparing the effectiveness and adverse effects of different systemic treatments for non-clear cell renal cell carcinoma. Eur Urol. 2017;71:426–436. doi:10.1016/j.eururo.2016.11.020
5. Chen W, Hill H, Christie A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539(7627):112–117. doi:10.1038/nature19796
6. Galandra C, Cerami C, Santi GC, et al. Job loss and health threatening events modulate risk-taking behaviours in the Covid-19 emergency. Sci Rep. 2020;10(1):22236. doi:10.1038/s41598-020-78992-x
7. Ning W, Lei S. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nature Biomedical Engineering. 2020;4:1197–1207. doi:10.1038/s41551-020-00633-5
8. Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, Evaluation, and Treatment of Coronavirus. Treasure Island (FL): StatPearls Publishing; 2020.
9. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi:10.1016/S0140-6736(20)30251-8
10. Li Y, Zhou W, Yang L, You R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol Res. 2020;157:104833. doi:10.1016/j.phrs.2020.104833
11. Hamming I, Cooper ME, Haagmans BL, et al. The emerging role of ACE2 in physiology and disease. J Pathol. 2007;212(1):1–11. doi:10.1002/path.2162
12. Sobczuk P, Szczylik C, Porta C, Czarnecka AM. Renin angiotensin system deregulation as renal cancer risk factor. Oncol Lett. 2017;14:5059–5068. doi:10.3892/ol.2017.6826
13. Larrinaga G, Pérez I, Sanz B, et al. Angiotensin-converting enzymes (ACE and ACE2) are downregulated in renal tumors. Regul Pept. 2010;165(2–3):218–223. doi:10.1016/j.regpep.2010.07.170
14. Mihalopoulos M, Dogra N, Mohamed N, Badani K, Kyprianou N. COVID-19 and kidney disease: molecular determinants and clinical implications in renal cancer. Eur Urol Focus. 2020;6(5):1086–1096. doi:10.1016/j.euf.2020.06.002
15. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi:10.1038/s41586-020-2286-9
16. Banerjee AK, Blanco MR, Bruce EA, et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–39.e21. doi:10.1016/j.cell.2020.10.004
17. Zhan W, Han T, Zhang C, et al. TRIM59 promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS One. 2015;10(11):e0142596. doi:10.1371/journal.pone.0142596
18. Hu YW, Kang CM, Zhao JJ, et al. LncRNA PLAC2 down-regulates RPL36 expression and blocks cell cycle progression in glioma through a mechanism involving STAT1. Journal of Cellular and Molecular Medicine. 2018;22(1):497–510. doi:10.1111/jcmm.13338
19. Shi XM, Zhao W, Yang YB, Lü BN. Expression of LOX in colorectal cancer tissues and its relationship with progress and prognosis. J Sichuan Univ Med Sci. 2017;48:566–569.
20. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary Oncol. 2015;19:A68–77. doi:10.5114/wo.2014.47136
21. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi:10.1093/bioinformatics/btp616
22. Zhou Y, Zhou B, Pache L, Chang M. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Int J Med. 2019;10:1523.
23. Peña-Llopis S, Brugarolas J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat Protoc. 2013;8(11):2240–2255. doi:10.1038/nprot.2013.141
24. von Roemeling CA, Radisky DC, Marlow LA, et al. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. 2014;74(17):4796–4810. doi:10.1158/0008-5472.CAN-14-0210
25. Wotschofsky Z, Gummlich L, Liep J, et al. Integrated microRNA and mRNA signature associated with the transition from the locally confined to the metastasized clear cell renal cell carcinoma exemplified by miR-146-5p. PLoS One. 2016;11(2):e0148746. doi:10.1371/journal.pone.0148746
26. Liep J, Kilic E, Meyer HA, Busch J, Jung K, Rabien A. Cooperative effect of miR-141-3p and miR-145-5p in the regulation of targets in clear cell renal cell carcinoma. PLoS One. 2016;11(6):e0157801. doi:10.1371/journal.pone.0157801
27. Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11(16):5730–5739. doi:10.1158/1078-0432.CCR-04-2225
28. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi:10.1016/j.neo.2017.05.002
29. Thul PJ, Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Science: a Publication of the Protein Society. 2018;27(1):233–244. doi:10.1002/pro.3307
30. Xu S, Xu H, Wang W, et al. The role of collagen in cancer: from bench to bedside. J translational med. 2019;17:309.
31. Walker C, Mojares E. Role of extracellular matrix in development and cancer progression. Int J Mol Sci. 2018;19.
32. Xu X, Jiang C, Wang S, et al. HPIP is upregulated in liver cancer and promotes hepatoma cell proliferation via activation of G2/M transition. IUBMB Life. 2013;65:873–882. doi:10.1002/iub.1202
33. Takeuchi H, Schneider M, Williamson DB, et al. Two novel protein O-glucosyltransferases that modify sites distinct from POGLUT1 and affect Notch trafficking and signaling. Proc Natl Acad Sci U S A. 2018;115:E8395–e402. doi:10.1073/pnas.1804005115
34. Majumder S, Crabtree JS. Targeting Notch in oncology: the path forward. Nat Rev Drug Discovery. 2020.
35. Schjoldager KT, Narimatsu Y. Global view of human protein glycosylation pathways and functions. Nature Reviews. Molecular Cell Biology. 2020;21(12):729–749. doi:10.1038/s41580-020-00294-x
36. He Q, Yang L, Gao K, et al. FTSJ1 regulates tRNA 2ʹ-O-methyladenosine modification and suppresses the malignancy of NSCLC via inhibiting DRAM1 expression. Nat Rev Drug Discovery. 2020;11:348.
37. Dewe JM, Fuller BL, Lentini JM, Kellner SM. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. The Journal of Pathology. 2017;212:37. doi:10.1128/MCB.00214-17
38. Wang B, Wan F, Sheng H, et al. Identification and validation of an 18-gene signature highly-predictive of bladder cancer metastasis. Sci Rep. 2018;8(1):374. doi:10.1038/s41598-017-18773-1
39. Zerdes I, Matikas A, Bergh J, Rassidakis GZ, Foukakis T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene. 2018;37:4639–4661. doi:10.1038/s41388-018-0303-3
40. Chevrier S, Levine JH, Zanotelli VRT, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169:736–49.e18. doi:10.1016/j.cell.2017.04.016
41. Clark DJ, Dhanasekaran SM, Petralia F, et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell. 2019;179:964–83.e31.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.