Back to Journals » OncoTargets and Therapy » Volume 16
JUND Promotes Tumorigenesis via Specifically Binding on Enhancers of Multiple Oncogenes in Cervical Cancer
Authors Zhou J, Mo J, Tan C, Xie F, Liang J, Huang W
Received 30 January 2023
Accepted for publication 13 May 2023
Published 31 May 2023 Volume 2023:16 Pages 347—357
DOI https://doi.org/10.2147/OTT.S405027
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Gaetano Romano
Jianlong Zhou,1,2,* Juanmei Mo,2,* Chaohui Tan,1,* Feng Xie,1 Jing Liang,3 Wenhua Huang1
1Xinhui People’s Hospital, Postdoctoral Innovation Practice Base of Southern Medical University, Xinhui, People’s Republic of China; 2Department of Oncology, Guangxi International Zhuang Medicine Hospital, Guangxi University of Chinese Medicine, Nanning, People’s Republic of China; 3The School of Basic Medical Science, Guangzhou University of Chinese Medicine, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wenhua Huang, School of Basic Medical Science, Southern Medical University, No. 1023 North Shatai Road, Guangzhou, 510515, People’s Republic of China, Email [email protected] Jing Liang, School of Basic Medical Science, Guangzhou University of Chinese Medicine, No. 232 East Waihuan Road, Guangzhou, 510006, People’s Republic of China, Email [email protected]
Purpose: Enhancers are genomic regulatory elements located distally from the target gene, which play a critical role in determining cell identity and function. Dysregulation of enhancers has been frequently observed in various types of cancer, including cervical cancer. However, the identity of enhancers and their associated transcriptional regulators that are involved in cervical cancer remains unclear.
Methods: With bioinformatics and 3D genomics, we revealed the enhancers in cervical cancer cell line and calculated which transcription factor (TF) is specifically binding on them based on TFs motif database. We knockdowned this TF and studied its function in cervical cancer cell line in vivo and in vitro.
Results: We found 14,826 activated enhancers and predicted that JUND (JunD Proto-Oncogene) is relatively enriched in the sequences of these enhancers. Well-known oncogene MYC and JUN were regulated by JUND through enhancers. To further explore the roles of JUND in cervical cancer, we analyzed the gene expression data of clinical cervical cancer samples and knock-downed JUND by CRISPR-Cas9 in Hela cell line. We found JUND is over-expressed in cervical cancer and the expression of JUND increased along with the cervical cancer progresses. Knockdown of JUND decreased the proliferation of Hela cells in vitro and in vivo and blocked cell cycle in G1-phase. Transcriptome sequencing analysis revealed the identification of 2,231 differentially expressed genes in response to the JUND knockdown treatment. This perturbation resulted in the modulation of several biological processes and pathways that have been previously linked to cancer.
Conclusion: These findings provide evidence for the significant involvement of JUND in cervical cancer pathogenesis, thereby positioning JUND as a potential therapeutic target for the treatment of this disease.
Keywords: cervical cancer, JUND, transcription factor, enhancer
Introduction
More than half a million women are diagnosed with cervical cancer every year and the disease caused over 300,000 deaths worldwide.1,2 Cervical cancer is always associated with infection of one or more of the oncogenic types of human papillomavirus (HPV).3 Without available HPV vaccination, cervical cancer is the second most common cancer among women in the third world countries. Even in developed countries, the prognosis of advanced/recurrent cervical cancer is still underwhelming and the lethality for cervical cancer is still rigorous.4 Cervical cancer cells show very different characteristics with normal cervical tissue, including the histological features, expression of genes and epigenetic modification of genome.5 Enhancers are genomic elements that bound by tissue- or cell type-specific transcription factors (TFs) that activate target gene transcription in a distal and autonomous manner.6 Redistribution of enhancers causes the disorder of gene expression in cancer.7 New cancer cell-specific enhancers are not shared by healthy cells from the same tissue.8,9 Enhancer specificity can extend to certain subtypes of cancer and be the specific targets for cancer therapy. With the epigenetic markers of histone H3 acetylated at lysine 27 (H3K27ac) and H3 monomethylated at K4 (H3K4me1) and enhancer–promoter interaction locations built by Paired-End-Tag sequencing (ChIA-PET),10–12 enhancers of cervical cancer can be found through integrated bioinformatic analysis.
TFs have been implicated as important drivers of cervical cancer.13,14 Certain TFs are identified to be lineage-specific and drive the cancer process through the activation of different enhancer repertoires.15,16 JUND, a member of the AP-1 family, functioned as an oncogene in cancers. It has been reported that JUND was a crucial modulator in prostate cell cycle progression and thus promoting cancer development.17 Ishikawa et al found that Butein ameliorated adult leukemia via inhibiting JUND expression.18 JUND was involved in cancer stemness and drug resistance in hepatocellular carcinoma.19 Previous study indicated that JUND is essential in prostate cancer proliferation and confer protection against radiation-induced cell death,20 while other AP-1 TFs family c-Jun and JunB had no effect on cell proliferation.21 It has been reported that the presence of HPV leads to an up-regulation of JUND expression in human ectocervical keratinocytes.22
In this study, we found the active enhancers in Hela cell line through bioinformatic. With the going deep of the research work, it had been found that JUND specifically binding on these enhancers. In order to study the tumorigenesis of JUND in cervical cancer, we decreased the expression of JUND through CRISPR-Cas9 genome editing tool in Hela cell line and investigated the cell proliferation in vivo. With the RNA-seq, the biological processes and pathways affected by JUND would be found. Our results demonstrated an essential role for JUND in cervical cancer initiation and tumorigenesis.
Materials and Methods
Cell Culture
The human Hela cell lines were obtained from ATCC (American Type Culture Collection). Hela cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, HyColne, Utah, USA) at 37°C in a humidified atmosphere containing 5% CO2.
Human Cancer Cell Xenograft Model
For tumor formation of Hela in NOD/SCID mice, 1.5×106 control Hela cells and 1.5×106 JUND-KO-pool Hela cells were harvested, washed twice with PBS, suspended in PBS with 30% Matrigel, and subcutaneously injected into hind legs of five NOD/SCID mice. The tumors were collected and weighted after 20 days. All animal experiments were approved by the Institutional Animal Care and Use Committee, with the the principles of the 3Rs (Replacement, Reduction and Refinement).
Genome Editing
For the knockdown of JUND in Hela, two expression cassettes encoding the sgRNA sequences (sgRNA1-JUND TCGGCGCGCAGGGGGGTAGG, sgRNA2-JUND CGGCGGGAAGGGCACAGGTT) flanking the deletion region were cloned into a plasmid that expresses a codon-optimized version of Cas9 (p2U6-Kana-Cas9-iRFP670). Seeding Hela cells to be 70% confluent in 6-well (5 × 105 cells) at transfection, 5 ug plasmid was transfected into the plasmid using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, USA) following the manufacturer’s guidelines. The transfected Hela cells were selected with puromycin for two days. We then used qPCR to test the mRNA expression of JUND and move on to the next steps.
Quantitative RT-PCR
Total RNA was purified from Hela with RNeasy Mini Kit (QIAGEN), and total RNA (1 µg) was reversely transcribed into cDNA and analyzed by qPCR. The primers for B-Actin are 5’-GCCAACACAGTGCTGTCT-3’ (forward) and 5’-AGGAGCAATGATCTTGATCTT-3’ (reverse). The primers for JUND are 5’-ATCGACATGGACACGCAGGAGC −3’ (forward) and 5’- CTCCGTGTTCTGACTCTTGAGG −3’ (reverse). Changes in mRNA expression were calculated according to the 2−ΔΔCT method (CT, cycle threshold).
Western Blot Analysis
The experiment involved extracting total proteins from cells using RIPA lysis buffer (Thermo Fisher Scientific) that contained a Protease and Phosphatase Inhibitor Cocktail (Cell Signaling Technology CST). The proteins were then separated using 10% SDS-PAGE and transferred to 0.45 µm nitrocellulose membranes (Merck Millipore USA). These membranes were then blocked using a blocking buffer and then incubated with primary antibodies overnight at 4°C. The primary antibodies used were anti-JUND (ab181615; Abcam) and anti-β-actin antibody (ab8226; Abcam). Afterward, the membranes were incubated with secondary antibodies for 1 hour at room temperature, and the detection of protein signals was done using chemiluminescence.
The Analysis of ChIA-PET and ChIP-Seq Data
The ChIA-PET and ChIP-seq data were obtained from Gene Expression Omnibus (GEO) and Encyclopedia of DNA Elements (ENCODE). The accessions were GSM1872889, GSM935328, GSM798322, GSM733696, GSM733684, GSM733682.23,24 The interaction data derived from POLII ChIA-PET analysis were displayed in BED12 format that showed the anchors and coordinates of the loop. The ChIP-Seq data were displayed using the UCSC Genome Browser (http://genome.ucsc.edu/). All analyses were performed using human (build hg19, GRCh37) RefSeq annotations downloaded from the UCSC genome browser.
Analysis of Gene Expression Profile
The gene expression profiles of cervical cancers, cervical intraepithelial neoplasia and normal cervical tissues were downloaded from GEO database (accession: GSE63514) (https://www.ncbi.nlm.nih.gov/geo/).25 The gene expression profiles of cervical cancer cell lines and normal cervical cells were downloaded from GEO database (accession: GSE89657).26 The cDNA of 24 normal, 14 cervical intraepithelial neoplasia grades 1 lesions, 22 cervical intraepithelial neoplasia grades 2 lesions, 40 cervical intraepithelial neoplasia grades 3 lesions, and 28 cancers specimens was hybridized to Affymetrix U133-Plus2.0 arrays. Differentially expressed gene among tumors, cervical intraepithelial neoplasia and normal tissues were identified using two-group comparisons, and p <0.05 was set as the cutoff criterion.
Analysis of Motif Enrichment
The AME tool in MEME suite is used to identify motifs that are relatively enriched in the sequences of AASEs with default parameter.27 The motif database is HOCOMOCO Human (v11 CORE)28.
RNA-Seq and Analysis
RNA purity was checked using the kaiaoK5500®Spectrophotometer (Kaiao, Beijing, China). RNA integrity and concentration were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A total amount of 2 μg RNA per sample was used as an input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (#E7530L, NEB, USA). Paired-end sequencing was completed on an Illumina HiSeq system. Clean data were generated after removing adapters and low quality reads. The reference GRCh38 genomes and the annotation file was downloaded from ENSEMBL database (http://www.ensembl.org/index.html). Bowtie2 v2.2.3 was used for building the genome index, and Clean Data was then aligned to the reference genome using HISAT2 v2.1.0. Reads Count for each gene in each sample was counted by Featurecounts.29 The differentially expressed genes were analyzed by DESeq2.30 The adjusted p-value <0.05 and Foldchange >2 as cutoff.
Biological Processes and Pathways Enrichment Analysis
We used DAVID (https://david.ncifcrf.gov/home.jsp.version6.8) to do KEGG pathway and GO analysis for the differentially expressed genes.31 EASE Score, a modified Fisher Exact P-value, is used to measure the gene-enrichment in annotation terms. P-value <0.05 to be considered strongly enriched in the annotation categories.
Cell Proliferation Assay
To assess cell proliferation, we employed the Cell Counting Kit 8 (CCK-8) assay following the manufacturer’s guidelines. Briefly, cells were seeded in 96-well plates at a density of 2×103 cells per well, and then treated with CCK-8 solution at various time points (24–96 hours) post-plating. The absorbance of the samples was measured at 450 nm using a microplate reader.
Cell Cycle Assay
To conduct the cell cycle assay, we employed the Cell Cycle Staining Kit (Multi Sciences Biotech Co., Ltd.) following the manufacturer’s instructions. Firstly, cells were harvested and treated with 100 μL RNase A at 37°C for 30 minutes and subsequently incubated with 400 μL Propidium iodide (PI) at 4°C in the dark for 30 minutes. Finally, we analyzed the cells using flow cytometry.
Statistics
Statistical analysis was carried out using GraphPad Prism 5. An unpaired two-tailed t-test was performed for comparisons between two groups. Results with p <0.05 were considered statistically significant. The mean values ± standard deviation (SD) were used to present the quantitative results.
Results
JUND Specially Bind on the Enhancers in Hela Cell Line
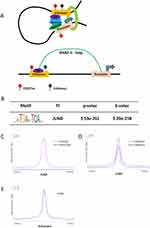
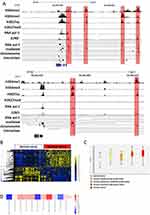
In order to find the enhancers involved in transcriptional regulation in cervical cancer, we analyzed the RNAPII ChIA-PET data of Hela cell line, which can help to detect promoter–enhancer interactions. Enhancers of Hela cell line were selected from the ends of RNAPII ChIA-PET loops with the mark of H3K27ac and H3K4me1 which acknowledged as the activated enhancer marker (Figure 1A) (Supplemental Table 1). Meanwhile, promoter regions were removed and we screened 14,826 activated enhancers. To find which TF plays a vital role in the formation of enhancer in Hela cell line. We analyzed the motifs of TFs which were enriched in the enhancers of Hela. In results, we found JUND was significantly enriched in them (Figure 1B). JUND is a member of the JUN family, and a functional component of the AP-1 TF complex. Results of distance analysis showed JUND specifically binding on H3K27ac, H3K4me1 and H3K4me3 area (Figure 1C and D). The peak of H3K4me1 was higher than H3K4me3, which means JUND prefer binding on the enhancers. Further, the peak of JUND located on the centre of activated enhancers in Hela cell lines (Figure 1E). We showed the genome regions of two well-known oncogenes MYC and JUN, of which enhancers have the peaks of JUND (Figure 2A).
JUND is Up-Regulated in Cervical Cancer
We analyzed the gene microarray data of 128 cervical tissue which includes normal, cervical cancer and cervical intraepithelial neoplasia.25 In our analysis, we verified the over-expression of JUND in cervical cancer (Figure 2B). Moreover, the expression of JUND increased along with the cancer progresses (Figure 2C). The expression of JUND of cervical cancer cell lines was higher compared to normal keratinocytes (Figure 2D). Given JUND’s specific binding to enhancers in Hela cells, we have concluded that it plays a critical role in the carcinogenic process by promoting the expression of oncogenes in cancer cells.
JUND is Required for the Proliferation of Hela Cell Line
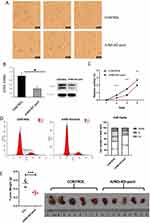
To prove that JUND is essential in cervical cancer, we knocked-out (KO) JUND in Hela cell line by CRISPR-Cas9 and collect the pools of resistance screened Hela cells (JUND-KO-pool) (Figure 3A). As results, the expression of JUND mRNA was decreased in JUND-KO-pool group, which has the same effect as knockdown (Figure 3B). The knockdown of JUND significantly reduced the proliferation of Hela cell line (Figure 3C). Meanwhile, cell cycle assay indicated that the cell number of JUND knockdown group in G1-phase was more than that of control group, and the cells of S-phage were decreased accordingly (Figure 3D). To explore the role of JUND in the tumorigenesis in vivo, the knockdown of JUND in Hela cells and controls were injected into the immunodeficient NOD-SCID mice, and the tumors of JUND knockdown group were smaller than control group (Figure 3E). These data demonstrated that JUND is required for the proliferation of Hela cells.
Multiple Processes and Pathways Were Affected by JUND
To reveal the progresses and pathways of JUND-dependent tumorigenesis of Hela cells, we performed RNA-seq analysis of control and JUND-KO pool of Hela cells to identify the genes that are affected by JUND. Over 2,231 genes were differentially expressed (adjusted p-value < 0.05 and Foldchange >2) between control and Hela cells with JUND knockdown (Figure 4A) (Supplemental Table 2). KEGG pathway enrichment analysis shows that differentially expressed genes were enriched in TGF-beta signaling pathway, ribosome, glycolysis/gluconeogenesis, extra cellular matrix (ECM)-receptor interaction and focal adhesion (Figure 4B). From the results of gene ontology (GO) enrichment analysis, we can find positive regulation of cell differentiation and regulation of programmed cell death enhanced (Figure 4C). However, cell cycle, positive relation of gene expression, intracellular transport, chromosome and mitochondrion organization were decreased (Figure 4C).
Discussion
Enhancers play a key role in tumorigenesis, and we intend to discover which TF specially regulates genes through enhancers in cervical cancer. With the RNAPII ChIA-PET data, H3K27ac and H3K4me1 ChIP-seq data, we found 14,826 activated enhancers in Hela cell line. Motif database was used to find the TF that specially binding on them. From the results, we found JUND (ATGACGTCAT) was significantly enriched on the activated enhancers in Hela cell line. With the analysis of the enhancer and promoter histone markers on the binding site of JUND, we found JUND prefer binding on the activated enhancers in Hela cell lines. JUND could promote the expression of target genes, which has been reported previously.21,32 In our results, the decrease of JUND led the down-regulated of proliferation and cell cycle was blocked in G1-phase. Therefore, the proportion of wild type Hela cells increased with time in the pool of JUND-KO group. The JUND-KO-pool cells proliferated fast after 3 days in vitro and the difference of tumors formed in vivo showed no excessively large.
From the RNA-seq data, we could find 2,231 differentially expressed genes and multiple processes and pathways were affected by JUND. The expression of MYC was effected by JUND through multiple enhancers. MYC has a central role in almost every aspect of the oncogenic process, orchestrating proliferation, apoptosis, differentiation, and metabolism.33 JUND probably activates the enhancers of MYC and promotes its transcription. Decrease of MYC could lead to the inhibition of cancer cells.34 From the results, it can be found another AP-1 TFs JUN is also regulated by JUND. AP-1 and MYC often collaborate to regulate the expression of genes which involved in cell cycle progression and proliferation.35,36 Researchers have shown that targeting both AP-1 and MYC together can be more effective than targeting either gene alone in certain types of cancer. This is because the two genes have overlapping functions and contribute to the same pathways that promote cancer growth and survival.37–39 Overexpression of JUND increases cell proliferation and invasiveness of cancer cell lines.40 The GO enrichment analysis shows positive regulation of cell differentiation and regulation of programmed cell death enhanced. Meanwhile, cell cycle, positive relation of gene expression, intracellular transport, chromosome and mitochondrion organization decreased. KEGG pathway enrichment analysis shows differentially expressed genes were enriched in TGF-beta signaling pathway, ribosome, glycolysis/gluconeogenesis, ECM–receptor interaction and focal adhesion.
Conclusion
Our research has validated the crucial role of JUND in the development of cervical cancer, as it facilitates gene transcription predominantly via enhancers. By examining the Hela cell line, we have identified a set of genes that are directly regulated by JUND. These findings offer a fresh perspective on the interplay between transcription factors and enhancers in the pathogenesis of cervical cancer.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval Statement
The animal experiments were approved by the Institutional Animal Care and Use Committee, with the principles of the 3Rs (Replacement, Reduction and Refinement).
Funding
This study is supported by the National Natural Science Foundation of China (Grant No. 82260784).
Disclosure
The authors declare that they have no competing interests.
References
1. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi:10.1016/S0140-6736(18)32470-X
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382(9895):889–899. doi:10.1016/S0140-6736(13)60022-7
4. Small WJ, Bacon MA, Bajaj A, et al. Cervical cancer: a global health crisis. Cancer-Am Cancer Soc. 2017;123(13):2404–2412.
5. Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: from pathogenesis to therapy. Tumour Biol. 2014;35(6):5083–5093. doi:10.1007/s13277-014-1737-z
6. Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–626. doi:10.1038/nrg3207
7. Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi:10.1016/j.cell.2013.09.053
8. Zhou J, Wang D, Tang D, Huang W. Abnormal activations of super-enhancers enhance the carcinogenicity in lung adenocarcinoma. Cancer Manag Res. 2020;12:8509–8518. doi:10.2147/CMAR.S258497
9. Hu CY, Mohtat D, Yu Y, et al. Kidney cancer is characterized by aberrant methylation of tissue-specific enhancers that are prognostic for overall survival. Clin Cancer Res. 2014;20(16):4349–4360. doi:10.1158/1078-0432.CCR-14-0494
10. Deng W, Blobel GA. Detecting long-range enhancer-promoter interactions by quantitative chromosome conformation capture. Methods Mol Biol. 2017;1468:51–62.
11. Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–837. doi:10.1016/j.molcel.2013.01.038
12. Barutcu AR, Fritz AJ, Zaidi SK, et al. C-ing the genome: a compendium of chromosome conformation capture methods to study higher-order chromatin organization. J Cell Physiol. 2016;231(1):31–35. doi:10.1002/jcp.25062
13. Wang C, Zou H, Chen A, et al. C-Myc-activated long non-coding RNA PVT1 enhances the proliferation of cervical cancer cells by sponging miR-486-3p. J Biochem. 2020;167(6):565–575. doi:10.1093/jb/mvaa005
14. Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–384. doi:10.1038/nature21386
15. Palstra RJ, Grosveld F. Transcription factor binding at enhancers: shaping a genomic regulatory landscape in flux. Front Genet. 2012;3:195. doi:10.3389/fgene.2012.00195
16. Hamdan FH, Johnsen SA. Perturbing enhancer activity in cancer therapy. Cancers. 2019;11:5. doi:10.3390/cancers11050634
17. Elliott B, Millena AC, Matyunina L, et al. Essential role of JunD in cell proliferation is mediated via MYC signaling in prostate cancer cells. Cancer Lett. 2019;448:155–167. doi:10.1016/j.canlet.2019.02.005
18. Ishikawa C, Senba M, Mori N. Butein inhibits NF-kappaB, AP-1 and Akt activation in adult T-cell leukemia/lymphoma. Int J Oncol. 2017;51(2):633–643. doi:10.3892/ijo.2017.4026
19. Cheng BY, Lau EY, Leung HW, et al. IRAK1 augments cancer stemness and drug resistance via the AP-1/AKR1B10 signaling cascade in hepatocellular carcinoma. Cancer Res. 2018;78(9):2332–2342. doi:10.1158/0008-5472.CAN-17-2445
20. Kajanne R, Miettinen P, Tenhunen M, Leppa S. Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. Int J Oncol. 2009;35(5):1175–1182. doi:10.3892/ijo_00000434
21. Millena AC, Vo BT, Khan SA. JunD is required for proliferation of prostate cancer cells and plays a role in transforming growth factor-beta (TGF-beta)-induced inhibition of cell proliferation. J Biol Chem. 2016;291(34):17964–17976. doi:10.1074/jbc.M116.714899
22. Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol. 2001;75(9):4283–4296. doi:10.1128/JVI.75.9.4283-4296.2001
23. Li G, Chen Y, Snyder MP, Zhang MQ. ChIA-PET2: a versatile and flexible pipeline for ChIA-PET data analysis. Nucleic Acids Res. 2017;45(1):e4. doi:10.1093/nar/gkw809
24. Pope BD, Ryba T, Dileep V, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515(7527):402–405. doi:10.1038/nature13986
25. den Boon JA, Pyeon D, Wang SS, et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A. 2015;112(25):E3255–E3264. doi:10.1073/pnas.1509322112
26. Marrero-Rodriguez D, la Cruz HA, Taniguchi-Ponciano K, et al. Kruppel like factors family expression in cervical cancer cells. Arch Med Res. 2017;48(4):314–322. doi:10.1016/j.arcmed.2017.06.011
27. Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi:10.1093/nar/gkp335
28. Kulakovskiy IV, Medvedeva YA, Schaefer U, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013;41:D195–D202. doi:10.1093/nar/gks1089
29. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi:10.1093/bioinformatics/btt656
30. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
31. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
32. Selvaraj N, Budka JA, Ferris MW, Plotnik JP, Hollenhorst PC. Extracellular signal-regulated kinase signaling regulates the opposing roles of JUN family transcription factors at ETS/AP-1 sites and in cell migration. Mol Cell Biol. 2015;35(1):88–100. doi:10.1128/MCB.00982-14
33. Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. doi:10.1038/s41392-018-0008-7
34. Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi:10.1016/j.cell.2012.03.003
35. Trop-Steinberg S, Azar Y. AP-1 expression and its clinical relevance in immune disorders and cancer. Am J Med Sci. 2017;353(5):474–483. doi:10.1016/j.amjms.2017.01.019
36. Alves DS, Mapekula L, Mdletshe N, Chetty D, Mowla S. HIV-1 transactivator of transcription (Tat) Co-operates With AP-1 factors to enhance c-MYC Transcription. Front Cell Dev Biol. 2021;9:693706. doi:10.3389/fcell.2021.693706
37. Wang C, Mayer JA, Mazumdar A, et al. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Mol Endocrinol. 2011;25(9):1527–1538. doi:10.1210/me.2011-1037
38. Lin SH, Wang HK, Yeh KT, et al. c-MYC expression in T (III/IV) stage oral squamous cell carcinoma (OSCC) patients. Cancer Manag Res. 2019;11:5163–5169. doi:10.2147/CMAR.S201943
39. Spender LC, Inman GJ. Developments in Burkitt’s lymphoma: novel cooperations in oncogenic MYC signaling. Cancer Manag Res. 2014;6:27–38. doi:10.2147/CMAR.S37745
40. Edwards J, Krishna NS, Mukherjee R, Bartlett JM. The role of c-Jun and c-Fos expression in androgen-independent prostate cancer. J Pathol. 2004;204(2):153–158. doi:10.1002/path.1605
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.