Back to Journals » Breast Cancer: Targets and Therapy » Volume 15
ITGB1BP1, a Novel Transcriptional Target of CD44-Downstream Signaling Promoting Cancer Cell Invasion
Authors Ahmad SMS, Nazar H, Rahman MM, Rusyniak RS, Ouhtit A
Received 31 January 2023
Accepted for publication 25 April 2023
Published 24 May 2023 Volume 2023:15 Pages 373—380
DOI https://doi.org/10.2147/BCTT.S404565
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Salma M S Ahmad, Hanan Nazar, Md Mizanur Rahman, Radoslaw Stefan Rusyniak, Allal Ouhtit
Biological Sciences Program, Department of Biological and Environmental Sciences, College of Arts and Science, Qatar University, Doha, Qatar
Correspondence: Allal Ouhtit, Department of Biological and Environmental Sciences, College of Arts and Science, Qatar University, PO Box 2713, Doha, Qatar, Tel +974 4403-7572, Email [email protected]
Abstract: Breast cancer (BC) is the most common malignancy worldwide and has a poor prognosis, because it begins in the breast and disseminates to lymph nodes and distant organs. While invading, BC cells acquire aggressive characteristics from the tumor microenvironment through several mechanisms. Thus, understanding the mechanisms underlying the process of BC cell invasion can pave the way towards the development of targeted therapeutics focused on metastasis. We have previously reported that the activation of CD44 receptor with its major ligand hyaluronan (HA) promotes BC metastasis to the liver in vivo. Next, a gene expression profiling microarray analysis was conducted to identify and validate CD44-downstream transcriptional targets mediating its pro-metastatic function from RNA samples collected from Tet CD44-induced versus control MCF7-B5 cells. We have already validated a number of novel CD44-target genes and published their underlying signaling pathways in promoting BC cell invasion. From the same microarray analysis, Integrin subunit beta 1 binding protein 1 (ITGB1BP1) was also identified as a potential CD44-target gene that was upregulated (2-fold) upon HA activation of CD44. This report will review the lines of evidence collected from the literature to support our hypothesis, and further discuss the possible mechanisms linking HA activation of CD44 to its novel potential transcriptional target ITGB1BP1.
Keywords: ITGB1BP1, Breast cancer, CD44, Hyaluronan, metastasis
Background
Breast cancer (BC) is the most common malignancy in women worldwide including Qatar.1,2 BC is a heterogeneous disease with altered biological and clinical characteristics.3 During tumor progression, cells undergo the process of epithelial-mesenchymal transition (EMT), triggering metastasis.3 Invasion is the recurring and defining event in the metastatic process,3 and elucidation of its mechanisms is critical for developing effective anti-metastatic therapies.
Invasion is a complex molecular network involving at least three major components, including cell adhesion molecules (CAMs)4,5 on the cell surface which facilitate the adhesion of invading cells to their surrounding extracellular matrix (ECM),6 proteinases that degrade the ECM, and growth factors that facilitate the growth of invading cells in a distant site. Cell adhesion maintains tissue structure and function, and changes in cell-cell and cell-matrix adhesion are of vital significance during invasion.5 Among the numerous CAM protein families, CD44 is the principal cell surface receptor for hyaluronic acid (HA), a major component of the ECM expressed by embryonic stem cells, connective tissue cells, bone marrow cells,7,8 and cancer cells.9,10 Binding of CD44 to HA stimulates conformational changes that triggers various oncogenic signaling pathways via various critical pathway networks (e.g., Rho GTPases, and PI3K/, AKT signaling pathways) leading to tumor cell survival, proliferation, and invasion.11
To better investigate the function of the standard form of CD44 (CD44s), in BC invasion/metastasis and further elucidate its downstream signaling, we have previously developed a tetracycline (Tet)-Off-regulated expression system of CD44s both in vitro12 and in vivo,13 and applied microarray analysis to identify several potential CD44s target genes. Based on functional annotations (cytoskeletal organization and motility, ECM degradation, cell survival, and cell growth), we have classified and validated three target genes along with their signaling pathways (Cortactin, Survivin and TGF-β2) as novel downstream target genes that underpin CD44-promoted breast tumor cell invasion.12,14,15
From the same microarray data, integrin subunit beta 1 binding protein 1 (ITGB1BP1) was selected for further validation studies as a potential target of CD44 because of its involvement in cell motility, metastasis, and integrin binding.
ITGB1BP1, also known as ICAP-1, binds to the cytoplasmic tail of β1 integrin.16 Specifically, it binds to the NPXY sequence motif found at the C-terminal of the β1 integrin through its C-terminal phosphotyrosine‐binding domain (PTB), which inhibits β1 integrin interaction with the ECM.16 Under normal circumstances, ITGB1BP1 plays a role in vascular differentiation,17 integrin activation, and focal adhesion (FA) formation.18 In this review, we collected and discussed data from the literature that support our hypothesis that ITGB1BP1 is a potential novel target of CD44-downstream signaling underlying the process of BC cell invasion.
Structure of ITGB1BP1
ITGB1BP1 is encoded by a gene located on the short arm of chromosome 2 (2p25.1), which produces two isoforms; a longer isoform (ITGB1BP1α), which is discussed here, and a shorter isoform (ITGB1BP1β), which lacks 50 C-terminal amino acids16 and is not well-studied. ITGB1BP1α has a molecular weight of 21,782 Da,16 and consists of two domains: a serine and a threonine-rich domain with a nuclear localization signal (NLS) sequence, as well as the PTB domain, which interacts with β1 integrin.16 The availability of these domains alternates based on ITGB1BP1’s conformational changes, allowing exposure of either NLS sequence or the integrin binding domain.16 For instance, when β1 integrin are overexpressed, the NLS sequence is masked, thus allowing ITGB1BP1 binding to β1 integrin and the localization of ITGB1BP1 in the cytoplasm.16 In eukaryotes, ITGB1BP1 is a phosphoprotein19 with multiple phosphorylation sites at the N-terminus as well as one site at the C-terminus.
The C-terminus features a protein kinase C phosphorylation site, while the N-terminal domain can be phosphorylated by protein kinase A (PKA), protein kinase C (PKC) and calcium/calmodulin-dependent protein kinase II (CaMKII) in order to regulate the biological activity of ITGB1BP1.20 Site-directed mutagenesis at Thr38 has shown that phosphorylation of ITGB1BP1 enhances cell spreading on a fibronectin matrix, while lack of phosphorylation at this site significantly inhibits cell spreading.20
Functions of ITGB1BP1
Physiologically, ITGB1BP1 is expressed in both normal and malignant cells. The following sections will discuss the role of ITGB1BP1 in both normal and malignant cells.
Physiological Functions of ITGB1BP1 in Normal Cells
ITGB1BP1 protein is present in all organs except the liver; however, ITGB1BP1 expression varies based on the tissue and cell type.19 While inhibition of ITG-β1 is lethal to embryogenesis, inhibition of ITGB1BP1 is on the contrary not lethal. In fact, previous studies have shown that mice lacking ITGB1BP1 were smaller and developed neurological disorders, bone defects,21 fertility defects and vascular defects.22 Moreover, ITGB1BP1 regulates osteoblast differentiation and proliferation.21 ITGB1BP1-deficient mice displayed retardation in growth and bone mineralization, and craniofacial deformity and absence of calvaria bone development, due to reduced cell proliferation and differentiation.21 Similar to results from in vivo studies21,23,24 in vitro experiments showed impairment in cell adhesion, and migration, and organization of fibronectin matrix in ITG1BP1-deficient osteoblasts.18 Furthermore, the inability of ITGB1BP1 to interact with mutant ITG-β1 also displayed similar abnormalities observed in ITGB1BP1-deficient osteoblasts, thus indicating that ITGB1BP1 is vital for osteoblast condensation, a significant and early step during differentiation.21
Physiological Functions of ITGB1BP1 in Cancer Cells
The following sections will discuss the role of ITGB1BP1 as a regulator of the mechanisms involved in cell proliferation, adhesion, and motility, processes involved in the onset and progression of cancer.
Physiological Functions of ITGB1BP1 in Cell Proliferation
ITGB1BP1 is known to interact specifically with the cytoplasmic domain of β1 integrin to control cell spreading on fibronectin matrix.19,25 Interestingly, ITGB1BP1 was not only observed in the cytoplasm but also in the nucleus, suggesting that it might act as a transcription factor.25 The transition of ITGB1BP1 between the nucleus and cytoplasm is β1 integrin dependent. While upregulated β1-integrin expression significantly inhibited ITGB1BP1 nuclear localization, this translocation to the nucleus is related to the stage of cell spreading on fibronectin;25 this suggests a role of ITGB1BP1 as a messenger that transmits information from integrin-dependent cell adhesion sites to the nucleus to regulate gene expression and cell proliferation.25 However, the underlying mechanisms of this phenomenon are still unclear. Nonetheless, while previous in vivo studies showed deregulation of cell proliferation in ITGB1BP1 deficient mice,21 overexpression of ITGB1BP1 in the nucleus was directly proportional to an increase in cell proliferation.25 Moreover, ITGB1BP1 induced cell proliferation in a fibronectin-dependent manner, possibly through the direct or indirect activation of the c-myc promoter and interaction with nuclear factors such as Nm23-H2.25 Nm23-H2 binds to a nuclease-hypersensitive element of the c-myc promoter, through which it activates ITGB1BP1-induced c-myc transcription and promotes cell proliferation along with upregulated cyclin D1 expression.25
Previous studies have indicated that integrin α5β1 interacts with receptor-tyrosine kinases and activates the ERK pathway, which is critical for cell proliferation. ERK pathway activation occurs through two key mechanisms associated with integrins. Integrins, through the cytoplasmic domain of their β subunit and the transmembrane segment of their α subunit, stimulate the Src family/focal adhesion kinase (FAK) pathway and the Shc/FAK pathway, respectively.26–29 The α subunit-dependent pathway enhances ERK activation.28 On the other hand, the β subunit-dependent pathway elongates ERK activation and promotes ERK nuclear translocation; this event is regulated by Rac.28 Moreover, β1 integrins also trigger the c-Jun NH2-terminal kinase signaling via the FAK/Cas/Rac pathway.30,31
Furthermore, ITGB1BP1 cooperates with Rho family GTPases, Rac and Cdc42, to regulate cell proliferation and cell motility.32 In fact, CD44 induced cell invasion via activation of RhoA GTPase/ROCK-1 signaling pathway.33 As mentioned above, ITGB1BP1 and Nm23-H2 regulate RhoA GTPase activity,25 suggesting that ITGB1BP1 and Nm23-H2 interaction can play a role in CD44-regulated tumor cell proliferation and invasion through the RhoA-GTPase pathway. CD44 is also involved in the activation of c-myc promoter; enhanced CD44 expression upregulates c-myc expression.34,35 In addition, CD44 also upregulates cyclin D1 by activating ERK pathway.36 ERK phosphorylation, triggers extracellular and intracellular signals to promote both cell proliferation and cell migration.37 The data suggests that ITGB1BP1 is linked to CD44-downstream signaling regulating cell proliferation and adhesion.
Physiological Functions of ITGB1BP1 in Cell Adhesion
Upon binding to their ligands, integrins merge into large clusters and recruit multiple proteins to form FAs to transduce signals to different subcellular compartments. FAs require Rho family GTPases, integrin engagement, and coordinated interaction between integrins and signaling molecules, as well as actin-binding proteins, actin microfilaments, and microtubules.21,38 Interestingly, Fournier et al created a double substitution of lysine for alanines in the NLS signal (KKNH)9 of ITGB1BP1, which abolished the function of NLS, subsequently leading to loss of cell adhesion.25 ITGB1BP1 protein, a negative regulator of cellular dissemination involves β1 integrin.32,38 Binding of ITGB1BP1 to β1 integrin adversely affects the integrin’s affinity for its ligand.21,24 Although a direct role of ITGB1BP1 is not known in FAs,38 loss of ITGB1BP1 results in the reorganization of FA in osteoblastic, fibroblastic and endothelial cells. Talin and kindlin bind the integrin’s cytoplasmic tail, and along with activated cytoskeletal and signaling proteins, they stimulate integrin binding to extracellular ligands.39 The PTB-domain of ITGB1BP1 attaches to kindlin-binding NPxY motif in β1 integrins and displaces inhibitory proteins, thus inhibiting talin-mediated integrin activation.23,40 Overexpression of ITGB1BP1 prevented talin-mediated β1 activation, leading to FA dissociation and subsequent loss of cell adhesion.23,38 Moreover, ITGB1BP1α, a β1A-integrin cytoplasmic partner, restricts the binding of both talin and kindlin to β1 integrin, thus preventing FA assembly.24 On the other hand, ITGB1BP1 impeded ROCK1-mediated cell contractility by regulating the affinity of β1 integrin;41 indicating transition of integrin between low and high affinity is necessary in regulating cell adhesion as well as the factors involved in maintaining an ECM environment.
Furthermore, CaMKII, a key regulator of ITGB1BP1α controls FA dynamics.42 CaMKII directly phosphorylates ITGB1BP1α and interrupts the intramolecular interaction between the N- and C-terminal domains of ITGB1BP1α; this exposes the PTB domain allowing binding of ITGB1BP1α to the β1 integrin tail and inhibits FA assembly.42 Overexpression of ITGB1BP1 increases CaMKII activity and decreases the FA size.42 In contrast, when ITGB1BP1 is inhibited, CaMKII does not interfere with FA assembly, suggesting that CaMKII acts on the β1 integrin-specific adhesion sites through interaction with ITGB1BP1, and subsequently promoting cell migration and destabilization of FAs.42 The increase in cytosolic calcium levels activates CaMKII pathway and controls HA synthesis as well as various signaling pathways, including MAPK pathway.43 On the other hand, increased HA synthesis promotes HA-CD44 binding, leading to the activation of various signaling pathways involved in the loss of cell-to-cell adhesion;9 this suggests an interaction between ITGB1BP1 and CD44 in regulating cell adhesion.
Physiological Functions of ITGB1BP1 in Cell Migration/Invasion
ITGB1BP1 forms a complex with ROCK-1 to promote cell migration via RhoA GTPase signaling pathway.44 ROCK-1 binds to ITGB1BP1 at N-terminal domain and PTB domain44 and overexpression of ITGB1BP1 recruits ROCK-1 allowing its translocation to the plasma membrane to form a complex with β1 integrin.44 RhoA induces membrane ruffles allowing its colocalization with β1 integrin.44 Cell migration and polarization depends mainly on the interaction between RhoA and ROCK-1,44 suggesting that ITGB1BP1 can enhance cell migration via activation of RhoA GTPase/ROCK pathway. Interestingly, CD44-HA interaction activates RhoA GTPase, leading to the recruitment of IP3 receptors, present in the intracellular calcium storage organelles, leading to calcium release into the cytoplasm;37 this results in CaMKII activation, followed by filamin phosphorylation and subsequent induction of tumor cell migration.37 Moreover, HA-CD44 binding phosphorylates myosin phosphatase and myosin light chain, leading to myosin adenosine triphosphatase activation to generate actomyosin-mediated cell migration.37 ROCK also phosphorylates NHE1 resulting in the alteration of ECM, thus inducing tumor cell migration and invasion.37 More interestingly, CD44 interacts with NHE1 to activate both hyaluronidase-2 and cathepsin, and promotes tumor cell invasion.45
Cell migration can be a result of another pathway that involves ubiquitylation of ITGB1BP1 by Smurf1 resulting in the transition from ROCK2-mediated to MRCKα-mediated cell contractility.18 In fact, HA-CD44 activates Cdc42 and phosphorylates PAK1 to form a complex with filamin and promotes cell migration and invasion.37 HA-CD44 can also activate Rac1 through the recruitment of ankyrin found in the cytosol, which interacts with Tiam1, leading to cancer cell progression.37 Activation of Rac1 also stimulates downstream effectors such as PAK and IQGAP1.46 IQGAPI-Cdc42 binding mediates various signaling events to activate actin cytoskeleton and tumor cell migration and invasion.47–49 Furthermore, IQGAPI complexes with ERK2 and MEK1/2 to activate ERK and MAPK signaling pathways, respectively, leading to tumor cell migration.50,51
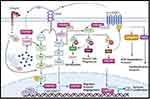
ITGB1BP1 also activates other oncogenic pathways by interacting with KRIT-1.22 KRIT-1 binds to ITGB1BP1 through its PTB domain, competing with β1 integrin to bind ITGB1BP1.52 Moreover, both ITGB1BP1 and KRIT-1 promote Notch signaling pathway leading to AKT phosphorylation and activation of PI3K/AKT pathway, which subsequently promote tumor cell survival and motility.53 As mentioned earlier, CD44 regulates PI3K/AKT pathway to induce tumor cell survival and motility,37 suggesting a plausible association between ITGB1BP1 and CD44. The data collected from the literature indicates that CD44 regulates ITGB1BP1 activation via various signaling pathways involved in mediating tumor cell invasion (Figure 1). Furthermore, bioinformatics analysis revealed several transcriptional factors including NRF1, HSF and ETS2, MZF1, ELK1 that promote the transcription of ITGB1BP1 due to the induction of PI3K/AKT and MAPK/ERK signaling pathways, respectively as shown in Figure 1.54 In fact, and as shown in Figure 1, CD44 interacts with its ligand HA and activates several oncogenic pathways. First, CD44 activates PI3K/AKT pathway, which is also activated by ITGB1BP1/KRT1 complex through phosphorylation of Notch1; This leads to the transcription of ITGB1BP1 by HSF and NRF1 transcription factors to enhance tumor cell migration and invasion. Activated PI3K/AKT pathway causes phosphorylation of Rho GEFs, which can also be activated by ITGB1BP1/Nm23-H2 complex. Activated Rho GEFs phosphorylate Rho GTPase, activating ROCK, and then ITGB1BP1, which translocate to the nucleus to form a complex with Nm23-H2 transcribing c-myc and cylinD1, thereby enhancing tumour cell proliferation and survival. On the other hand, activated ROCK may also phosphorylate NHE1 to trigger the expression of HA through Hyal-2, as well as the expression of MMP9, leading to increased tumor cell invasion. In addition, activated Rho GEFs may also phosphorylate Cdc42 GTPase, activating PAK IQGAP1 leading to an increase in tumor cell migration. Interestingly, CD44 can trigger the transcription of its target ITGB1BP1 by activating MZF1, ETS2, and ELK1 transcription factors via the MEK/ERK pathway, to promote tumor cell migration and invasion.
Conclusion
Our review has provided several lines of evidence, supporting our hypothesis that CD44-HA interaction would induce various oncogenic intermediate signaling pathways, which in turn release various transcription factors that lead to the transactivation of the CD44-target, ITGB1BP1.In fact, CD44 activates the transcription of ITGB1BP1 at least via PI3K/AKT, MAPK/ERK signaling pathways, which supports our hypothesis that ITGB1BP1 is a downstream potential novel transcriptional target of CD44/HA promoting tumor cell invasion and metastasis.
Abbreviations
AKT, Protein kinase B; BC, Breast cancer; CAM, Cell adhesion molecule; CaMKII, Calcium/calmodulin-dependent protein kinase II; Cas, CRISPR-associated proteins; CD44, Cluster of differentiation 44; Cdc42, Cell division control protein 42 homolog; ECM, Extracellular matrix; EMT, Epithelial-mesenchymal transition; ERK, Extracellular-signal-regulated kinase; FAs, Focal adhesions; FAK, Focal adhesion kinase; HA, Hyaluronic acid; ICAP-1, Integrin cytoplasmic-associated protein 1; ITG-β1, Integrin subunit beta-1; ITGB1BP1, Integrin Subunit Beta 1 Binding Protein 1; KRIT-1, Krev interaction trapped protein 1; NLS, Nuclear localization signal; Nm23-H2, Nucleoside diphosphate kinase B; PI3K, phosphoinositide 3-kinase; PAK, p21-activated kinases; PK, Protein kinase; PTB, Phosphotyrosine‐binding domain; Rac1, Ras-related C3 botulinum toxin substrate 1; Rho, Ras homologous; ROCK, Rho-associated protein kinase; Smurf1, Smad, ubiquitin regulatory factor 1; Tet, Tetracycline; TGF-β2, Transforming growth factor beta 2.
Consent for Publication
Yes.
Acknowledgments
We are grateful to Dr Ishita Gupta for minor contribution to this manuscript.
Funding
This research was funded by Qatar University (Internal grant number (QUST-1-CAS2019-22)), and the Qatar National Research Foundation: UREP24-117-1-027 and UREP29-186-3-059. Dr Hanan Nazar was awarded a PhD scholarship from the Kuwaiti Government.
Disclosure
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript, or in the decision to publish the results.
References
1. Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi:10.1002/ijc.31937
2. Narayan AK, Al-Naemi H, Aly A, et al. Breast cancer detection in Qatar: evaluation of mammography image quality using a standardized assessment tool. Eur J Breast Health. 2020;16(2):124–128. doi:10.5152/ejbh.2020.5115
3. McSherry EA, Donatello S, Hopkins AM, McDonnell S. Molecular basis of invasion in breast cancer. Cell Mol Life Sci. 2007;64(24):3201–3218. doi:10.1007/s00018-007-7388-0
4. Martin T, Ye L, Sanders AJ, Lane J, Jiang WG. Cancer invasion and metastasis: molecular and cellular perspective. Jandial R, editor. In: Madame Curie Bioscience Database. Landes Bioscience; 2013. Available from https://www.ncbi.nlm.nih.gov/books/NBK164700/.
5. Martin TA, Jiang WG. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta. 2009;1788(4):872–891. doi:10.1016/j.bbamem.2008.11.005
6. Bendas G, Borsig L. Cancer cell adhesion and metastasis: selectins, integrins, and the inhibitory potential of heparins. Int J Cell Biol. 2012;2012:676731. doi:10.1155/2012/676731
7. Domev H, Amit M, Laevsky I, Dar A, Itskovitz-Eldor J. Efficient engineering of vascularized ectopic bone from human embryonic stem cell-derived mesenchymal stem cells. Tissue Eng Part A. 2012;18(21–22):2290–2302. doi:10.1089/ten.TEA.2011.0371
8. Gronthos S, Franklin DM, Leddy HA, Robey PG, Storms RW, Gimble JM. Surface protein characterization of human adipose tissue-derived stromal cells. J Cell Physiol. 2001;189(1):54–63. doi:10.1002/jcp.1138
9. Isacke CM, Yarwood H. The hyaluronan receptor, CD44. Int J Biochem Cell Biol. 2002;34(7):718–721. doi:10.1016/s1357-2725(01)00166-2
10. Lee JY, Spicer AP. Hyaluronan: a multifunctional, megaDalton, stealth molecule. Curr Opin Cell Biol. 2000;12(5):581–586. doi:10.1016/s0955-0674(00)00135-6
11. Naor D, Nedvetzki S, Golan I, Melnik L, Faitelson Y. CD44 in cancer. Crit Rev Clin Lab Sci. 2002;39(6):527–579. doi:10.1080/10408360290795574
12. Hill A, McFarlane S, Mulligan K, et al. Cortactin underpins CD44-promoted invasion and adhesion of breast cancer cells to bone marrow endothelial cells. Oncogene. 2006;25(45):6079–6091. doi:10.1038/sj.onc.1209628
13. Ouhtit A, Abd Elmageed ZY, Abdraboh ME, Lioe TF, Raj MHG. In vivo evidence for the role of CD44s in promoting breast cancer metastasis to the liver. Am J Pathol. 2007;171(6):2033–2039. doi:10.2353/ajpath.2007.070535
14. Abdraboh ME, Gaur RL, Hollenbach AD, Sandquist D, Raj MH, Ouhtit A. Survivin is a novel target of CD44-promoted breast tumor invasion. Am J Pathol. 2011;179(2):555–563. doi:10.1016/j.ajpath.2011.04.042
15. Ouhtit A, Madani S, Gupta I, et al. TGF-beta2: a novel target of CD44-promoted breast cancer invasion. research paper. J Cancer. 2013;4(7):566–572. doi:10.7150/jca.6638
16. Chang DD, Wong C, Smith H, Liu J. ICAP-1, a novel beta1 integrin cytoplasmic domain-associated protein, binds to a conserved and functionally important NPXY sequence motif of beta1 integrin. J Cell Biol. 1997;138(5):1149–1157. doi:10.1083/jcb.138.5.1149
17. Brütsch R, Liebler SS, Wüstehube J, et al. Integrin cytoplasmic domain-associated protein-1 attenuates sprouting angiogenesis. Circ Res. 2010;107(5):592–601. doi:10.1161/circresaha.110.217257
18. Bouin AP, Kyumurkov A, Régent-Kloeckner M, et al. ICAP-1 monoubiquitylation coordinates matrix density and rigidity sensing for cell migration through ROCK2-MRCKα balance. J Cell Sci. 2017;130(3):626–636. doi:10.1242/jcs.200139
19. Zhang XA, Hemler ME. Interaction of the integrin β1 cytoplasmic domain with ICAP-1 Protein. J Biol Chem. 1999;274(1):11–19. doi:10.1074/jbc.274.1.11
20. Bouvard D, Block MR. Calcium/calmodulin-dependent protein kinase II controls integrin α5β1-mediated cell adhesion through the integrin cytoplasmic domain associated protein-1α. Biochem Biophys Res Commun. 1998;252(1):46–50. doi:10.1006/bbrc.1998.9592
21. Bouvard D, Aszodi A, Kostka G, Block MR, Albigès-Rizo C, Fässler R. Defective osteoblast function in ICAP-1-deficient mice. Development. 2007;134(14):2615–2625. doi:10.1242/dev.000877
22. Faurobert E, Rome C, Lisowska J, et al. CCM1-ICAP-1 complex controls β1 integrin-dependent endothelial contractility and fibronectin remodeling. J Cell Biol. 2013;202(3):545–561. doi:10.1083/jcb.201303044
23. Brunner M, Millon-Frémillon A, Chevalier G, et al. Osteoblast mineralization requires beta1 integrin/ICAP-1-dependent fibronectin deposition. J Cell Biol. 2011;194(2):307–322. doi:10.1083/jcb.201007108
24. Millon-Frémillon A, Bouvard D, Grichine A, Manet-Dupé S, Block MR, Albiges-Rizo C. Cell adaptive response to extracellular matrix density is controlled by ICAP-1-dependent beta1-integrin affinity. J Cell Biol. 2008;180(2):427–441. doi:10.1083/jcb.200707142
25. Fournier H-N, Dupé-Manet S, Bouvard D, et al. Nuclear translocation of integrin cytoplasmic domain-associated protein 1 stimulates cellular proliferation. Mol Biol Cell. 2005;16(4):1859–1871. doi:10.1091/mbc.e04-08-0744
26. Wary KK, Mainiero F, Isakoff SJ, Marcantonio EE, Giancotti FG. The adaptor protein Shc couples a class of integrins to the control of cell cycle progression. Cell. 1996;87(4):733–743. doi:10.1016/s0092-8674(00)81392-6
27. Pozzi A, Wary KK, Giancotti FG, Gardner HA. Integrin alpha1beta1 mediates a unique collagen-dependent proliferation pathway in vivo. J Cell Biol. 1998;142(2):587–594. doi:10.1083/jcb.142.2.587
28. Hirsch E, Barberis L, Brancaccio M, et al. Defective Rac-mediated proliferation and survival after targeted mutation of the beta1 integrin cytodomain. J Cell Biol. 2002;157(3):481–492. doi:10.1083/jcb.200111065
29. Barberis L, Wary KK, Fiucci G, et al. Distinct roles of the adaptor protein Shc and focal adhesion kinase in integrin signaling to ERK. J Biol Chem. 2000;275(47):36532–36540. doi:10.1074/jbc.M002487200
30. Dolfi F, Garcia-Guzman M, Ojaniemi M, Nakamura H, Matsuda M, Vuori K. The adaptor protein Crk connects multiple cellular stimuli to the JNK signaling pathway. Proc Natl Acad Sci U S A. 1998;95(26):15394–15399. doi:10.1073/pnas.95.26.15394
31. Oktay M, Wary KK, Dans M, Birge RB, Giancotti FG. Integrin-mediated activation of focal adhesion kinase is required for signaling to Jun NH2-terminal kinase and progression through the G1 phase of the cell cycle. J Cell Biol. 1999;145(7):1461–1469. doi:10.1083/jcb.145.7.1461
32. Degani S, Balzac F, Brancaccio M, et al. The integrin cytoplasmic domain-associated protein ICAP-1 binds and regulates Rho family GTPases during cell spreading. J Cell Biol. 2002;156(2):377–387. doi:10.1083/jcb.200108030
33. Aubert L, Guilbert M, Corbet C, et al. NGF-induced TrkA/CD44 association is involved in tumor aggressiveness and resistance to lestaurtinib. Oncotarget. 2015;6(12):9807–9819. doi:10.18632/oncotarget.3227
34. Chen P-C, Yu -C-C, Huang W-Y, et al. c-Myc acts as a competing endogenous RNA to Sponge miR-34a, in the upregulation of CD44, in urothelial carcinoma. Cancers. 2019;11(10):1457. doi:10.3390/cancers11101457
35. Park J, Kim SY, Kim H-J, Kim K-M, Choi EY, Kang M-S. A reciprocal regulatory circuit between CD44 and FGFR2 via c-myc controls gastric cancer cell growth. Oncotarget. 2016;7(19):28670–28683. doi:10.18632/oncotarget.8764
36. Kothapalli D, Flowers J, Xu T, Puré E, Assoian RK. Differential activation of ERK and Rac mediates the proliferative and anti-proliferative effects of hyaluronan and CD44. J Biol Chem. 2008;283(46):31823–31829. doi:10.1074/jbc.M802934200
37. Bourguignon LYW. Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin Cancer Biol. 2008;18(4):251–259. doi:10.1016/j.semcancer.2008.03.007
38. Bouvard D, Vignoud L, Dupé-Manet S, et al. Disruption of focal adhesions by integrin cytoplasmic domain-associated protein-1α. J Biol Chem. 2003;278(8):6567–6574. doi:10.1074/jbc.M211258200
39. Calderwood DA, Campbell ID, Critchley DR. Talins and kindlins: partners in integrin-mediated adhesion. Nat Rev Mol Cell Biol. 2013;14(8):503–517. doi:10.1038/nrm3624
40. Pouwels J, Nevo J, Pellinen T, Ylänne J, Ivaska J. Negative regulators of integrin activity. J Cell Sci. 2012;125(Pt 14):3271–3280. doi:10.1242/jcs.093641
41. Faurobert E, Albiges-Rizo C. Recent insights into cerebral cavernous malformations: a complex jigsaw puzzle under construction. Febs J. 2010;277(5):1084–1096. doi:10.1111/j.1742-4658.2009.07537.x
42. Millon-Frémillon A, Brunner M, Abed N, et al. Calcium and calmodulin-dependent serine/threonine protein kinase type II (CaMKII)-mediated intramolecular opening of integrin cytoplasmic domain-associated protein-1 (ICAP-1α) negatively regulates β1 integrins. J Biol Chem. 2013;288(28):20248–20260. doi:10.1074/jbc.M113.455956
43. Rauhala L, Hämäläinen L, Salonen P, et al. Low dose ultraviolet B irradiation increases hyaluronan synthesis in epidermal keratinocytes via sequential induction of hyaluronan synthases Has1-3 mediated by p38 and Ca2+/calmodulin-dependent protein kinase II (CaMKII) signaling. J Biol Chem. 2013;288(25):17999–18012. doi:10.1074/jbc.M113.472530
44. Stroeken PJM, Alvarez B, Van Rheenen J, et al. Integrin cytoplasmic domain-associated protein-1 (ICAP-1) interacts with the ROCK-I kinase at the plasma membrane. J Cell Physiol. 2006;208(3):620–628. doi:10.1002/jcp.20699
45. Bourguignon LYW, Singleton PA, Diedrich F, Stern R, Gilad E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J Biol Chem. 2004;279(26):26991–27007. doi:10.1074/jbc.m311838200
46. Ruiz-Velasco R, Lanning CC, Williams CL. The activation of Rac1 by M3 muscarinic acetylcholine receptors involves the translocation of Rac1 and IQGAP1 to cell junctions and changes in the composition of protein complexes containing Rac1, IQGAP1, and actin. J Biol Chem. 2002;277(36):33081–33091. doi:10.1074/jbc.M202664200
47. Fukata M, Kuroda S, Nakagawa M, et al. Cdc42 and Rac1 regulate the interaction of IQGAP1 with beta-catenin. J Biol Chem. 1999;274(37):26044–26050. doi:10.1074/jbc.274.37.26044
48. Ho YD, Joyal JL, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem. 1999;274(1):464–470. doi:10.1074/jbc.274.1.464
49. Kuroda S, Fukata M, Nakagawa M, et al. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin- mediated cell-cell adhesion. Science. 1998;281(5378):832–835. doi:10.1126/science.281.5378.832
50. Ren JG, Li Z, Sacks DB. IQGAP1 modulates activation of B-Raf. Proc Natl Acad Sci U S A. 2007;104(25):10465–10469. doi:10.1073/pnas.0611308104
51. Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol Cell Biol. 2005;25(18):7940–7952. doi:10.1128/mcb.25.18.7940-7952.2005
52. Zheng Y, Qiu J, Hu J, Wang G. Concepts and hypothesis: integrin cytoplasmic domain-associated protein-1 (ICAP-1) as a potential player in cerebral cavernous malformation. J Neurol. 2013;260(1):10–19. doi:10.1007/s00415-012-6567-6
53. Guo S, Liu M, Gonzalez-Perez RR. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochimica Et Biophysica Acta. 2011;1815(2):197–213. doi:10.1016/j.bbcan.2010.12.002
54. Daily K, Patel VR, Rigor P, Xie X, Baldi P. MotifMap: integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinform. 2011;12:495. doi:10.1186/1471-2105-12-495
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.