Back to Journals » Infection and Drug Resistance » Volume 15
IS26-Mediated Formation of a Hybrid Plasmid Carrying mcr-1.1
Authors Wu R, Lv L, Wang C, Gao G, Yu K, Cai Z, Liu Y, Yang J, Liu JH
Received 28 September 2022
Accepted for publication 23 November 2022
Published 9 December 2022 Volume 2022:15 Pages 7227—7234
DOI https://doi.org/10.2147/IDR.S390765
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Renjie Wu,1– 3,* Luchao Lv,1– 3,* Chengzhen Wang,1– 3 Guolong Gao,1– 3 Kaiyang Yu,1– 3 Zhongpeng Cai,1– 3 Yiyun Liu,1– 3 Jun Yang,1– 3 Jian-Hua Liu1– 4
1Key Laboratory of Zoonosis of Ministry of Agricultural and Rural Affairs, College of Veterinary Medicine, South China Agricultural University, Guangzhou, People’s Republic of China; 2Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, College of Veterinary Medicine, South China Agricultural University, Guangzhou, People’s Republic of China; 3National Risk Assessment Laboratory for Antimicrobial Resistance of Microorganisms in Animal, College of Veterinary Medicine, South China Agricultural University, Guangzhou, People’s Republic of China; 4Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jian-Hua Liu, College of Veterinary Medicine, South China Agricultural University, Wushan Road 483, Guangzhou, 510642, People’s Republic of China, Email [email protected]
Purpose: The objective of this study was to elucidate the characteristics and mechanism of formation of the fusion plasmid pHNSHP24 carrying mcr-1.1.
Materials and Methods: mcr-1.1-bearing Escherichia coli SHP24 and the corresponding transconjugant were subjected to whole-genome sequencing (WGS) combining the Illumina and MinION platforms to obtain the complete sequences of the fusion plasmid and its original plasmids.
Results: Complete sequence analysis and S1 nuclease-pulsed field gel electrophoresis (S1-PFGE) results indicated that E. coli SHP24 carried four plasmids: mcr-1.1-harboring phage-like plasmid pHNSHP24-3, F53:A-:B- plasmid pHNSHP24-4, pHNSHP24-1, and pHNSHP24-2. However, the plasmid pHNSHP24 carrying mcr-1.1 presents in the transconjugant differed from the four plasmids in the donor strain SHP24. Further analysis showed that pHNSHP24 may be the fusion product of pHNSHP24-3 and pHNSHP24-4 and is formed through a replicative transposition mechanism mediated by IS 26 in E. coli SHP24.
Conclusion: This study is the first to report the fusion of an mcr-1.1-harboring phage-like pO111 plasmid and an F53:A-:B- plasmid mediated by IS 26. Our findings revealed the role of phage-like and fusion plasmids in the dissemination of mcr-1.1.
Keywords: fusion plasmid, mcr-1.1, IS 26, pO111 plasmid
Introduction
The emergence and global transmission of the plasmid-mediated colistin resistance gene mcr-1.1 poses a great threat to public health.1,2 More than ten types of mcr-1.1-carrying plasmids have been reported, with IncI2, IncX4, and IncHI2 plasmids being the dominant vehicles that mediate the spread of mcr-1.1 among different Enterobacteriaceae species from various sources worldwide. mcr-1.1-bearing phage-like pO111 plasmids have also been reported in animal, human, or environmental original isolates from China and have gradually become an important vehicle that mediates the dissemination of mcr-1.1.3–5 The original pO111 plasmid pO111_2 was identified as a bacteriophage P1-like plasmid and found in enterohemorrhagic Escherichia coli of the O111:H1 serotype in Japan in 2001.6 Thereafter, phage-like pO111 plasmids were isolated from E. coli, Klebsiella pneumoniae, and Salmonella enterica.7–9 In addition to the mcr-1.1 gene, phage-like pO111 plasmids also carried other resistance genes, such as aadA, oqxAB, floR, blaCTX-M, blaTEM and so on.8,10,11 Similar to other phage-like plasmids, pO111 plasmids lack conjugative transfer ability.7–9,12
However, pO111 plasmids can be inserted into conjugative plasmids, such as IncF33:A-:B-, and IncFIA, and thereby obtaining conjugation capabilities.7,12 Furthermore, an increasing number of hybrid plasmids have been reported in recent years. Some hybrid plasmids are formed through recombination mediated by insertion sequences, transposons, or other homologous regions, such as IS26, IS1294, IS1216E, and Tn1721.7,13–16 Some hybrid plasmids are formed through multiple homologous recombinations, such as the IncX3-X4 hybrid plasmid pCQ02-121. The plasmid pCQ02-121 is formed through two recombinations mediated by both IS26 and nic site, which is a specific site cleaved by relaxation enzymes on the origin of transfer.17,18
Here, we characterized an mcr-1.1-carrying hybrid plasmid formed through the fusion of IncF53:A-:B- and phage-like pO111 plasmids and depicted the possible fusion mechanism.
Materials and Methods
Bacterial Strain
The mcr-1.1-positive E. coli strain SHP24 and its transconjugant C600/pHNSHP24 as well as its transformant E. coli DH5α/pHNSHP24 carrying mcr-1.1 were obtained in our previous study.5
S1 Nuclease-Pulsed Field Gel Electrophoresis (S1-PFGE)
The plasmid profiles of SHP24 and its transconjugant were determined using S1-PFGE. Briefly, the DNA of SHP24 and its transconjugant were digested with S1 nuclease (Takara, Japan), and the DNA of the S. enterica serotype Braenderuo H9812 was digested with XbaI restriction enzymes (Takara, Japan). The digested DNA was subjected to S1-PFGE using the CHEF-MAPPER System (Bio-Rad, USA), and the results were visualized using a gel imaging system (Bio-Rad, USA).
Whole-Genome Sequencing (WGS) Analysis
E. coli SHP24 and its transconjugant E. coli C600/pHNSHP24 were subjected to WGS combining the Illumina HiSeq and Oxford Nanopore MinION platforms, and complete sequences were generated through de novo hybrid assembly using Unicycler version 0.4.3.19 Multilocus sequence types (MLST), antimicrobial resistance genes, plasmid replicon types, and plasmid MLST (pMLST) were analyzed using the CGE server (https://cge.cbs.dtu.dk/services/). Insertion sequence (IS) elements were identified using the ISfinder (https://isfinder.biotoul.fr/). The phage sequence was identified using PHASTER.20 The plasmid sequences were annotated using the RAST server (https://rast.nmpdr.org/). The complete sequence comparison of plasmids was performed using Easyfig.21
Analysis of mcr-1.1-Bearing Plasmids in the NCBI Database
In the NCBI database, we used the complete sequence of mcr-1.1 to search for the complete sequences of mcr-1.1-bearing plasmids, and a total of 627 complete sequences of mcr-1.1-bearing plasmids were obtained. Antibiotic resistance genes and replicon types of mcr-1.1-carrying plasmids were retrieved using ABRicate (https://github.com/tseemann/abricate). Information on these plasmids was summarized and statistically analyzed (Tables S1–S3).
Conjugation Frequencies of Fusion Plasmid
The conjugation frequencies of pHNSHP24 were determined by conjugation assays using strain SHP24 or the transformant E. coli DH5α/pHNSHP24 as the donors and streptomycin-resistant E. coli C600 as the recipient. The experiment was carried out with biological triplicate. Conjugation frequencies were calculated as the number of transconjugants per recipient. Transconjugants were selected on lysogeny broth (LB) agar plates supplemented with colistin (2 µg/mL) and streptomycin (3000 µg/mL). PCR was performed to confirm the presence of mcr-1 and the corresponding replicon type in the transconjugants (Table S4). In addition, PCR was performed to detect the presence of the fusion plasmid pHNSHP24 in parental strain SHP24 by using two pairs of specific primers Hybrid-1/2-F/R (Table S4).
Nucleotide Sequence Accession Number
The complete nucleotide sequence of the fusion pHNSHP24 was deposited in GenBank under the accession number CP065023.
Results
Identification of the Fusion Plasmid in the Transconjugant C600/pHNSHP24
The mcr-1.1-bearing plasmid pHNSHP24 in transconjugant belonged to the IncFII group which was rarely reported to carry the mcr-1.1 gene.5 Therefore, we sequenced pHNSHP24 to study the characteristics of this plasmid. Sequence analysis showed that pHNSHP24 is a 174812-bp hybrid plasmid harboring pO111 and F53:A-:B- plasmid replicons (Table 1). However, the S1-PFGE results showed that the size of the plasmid pHNSHP24 carried by the transconjugant was different from that of the plasmids carried by the donor SHP24 (Figure S1).
 |
Table 1 Characterization of E. coli Strain SHP24 and Its Transconjugant Used in This Study |
Analysis of Plasmids in SHP24
To further explore the source of plasmid pHNSHP24, the complete sequence of SHP24 was also obtained using WGS combining the Illumina and MinION platforms. Sequence analysis showed that the E. coli strain SHP24 belonged to ST6913 and harbored four plasmids, namely pHNSHP24-1 (IncHI2, 259,017 bp), pHNSHP24-2 (IncFII(pCoo), 186,621 bp), pHNSHP24-3 (pO111, 99,354 bp), and pHNSHP24-4 (IncF53:A-:B-; 76,121 bp). The IncHI2 plasmid pHNSHP24-1 harbored the resistance genes aac(3)-IV, aac(6’)-Ib-cr, aadA1, aadA2b, aph(3’)-Ia, aph(4)-Ia, arr-3, blaOXA-1, catB3, cmlA1, dfrA12, floR, oqxAB, sul1, sul2, and sul3. The InFII(pCoo) plasmid pHNSHP24-2 and IncF53:A-:B- plasmid pHNSHP24-4 both harbored no resistance genes, and the colistin resistance gene mcr-1.1 was carried by the phage-like pO111 plasmid pHNSHP24-3 (Table 1). Although the WGS data of the original strain SHP24 lack the sequence of the hybrid plasmid, we detected it in SHP24 by PCR.
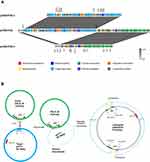
The pO111 plasmid pHNSHP24-3 encodes 159 predicted open reading frames (ORFs), most of which are related to phage-related genes. Plasmid pHNSHP24-3 exhibited high homology to Escherichia phage P1 (AF234172) and P7 (AF503408), mcr-1.1-positive phage-like IncY plasmid pHYEC7-mcr1 (KX518745), mcr-1.1-carrying phage-like pO111 plasmids pPC6-mcr1 (CP080256) and pMCR_SCKP-LL83 (MF510496), which originated from E. coli and K. pneumoniae in China, with 97–99% identity and 69–92% coverage (Figure 1).
The F53:A-:B- plasmid pHNSHP24-4 had a typical IncFII plasmid conjugative transfer region structure, including tra, trb, and finO genes, and exhibited high homology to the mcr-1.1-bearing IncFII plasmid pCAU16175_3 (CP047381) and floR-harboring IncFII plasmid pCP8-3-IncFII (CP053737), originating from E. coli in China, with 94–99% identity and 68–77% coverage (Figure 1).
Sequence Comparison and Proposed Formation Mechanism of the Fusion Plasmid pHNSHP24
The complete sequence of pHNSHP24 was 174,812 bp with a G+C content of 49%, and only harbored the colistin resistance gene mcr-1.1. BLASTn results showed that pHNSHP24 was highly similar to the mcr-1.1-bearing pO111-IncN-IncF33:A-:B- plasmid pD72C (MK419152) with 81% coverage and 98% identity (Figure 1). Further analysis indicated that the fusion plasmid pHNSHP24 consisted of pHNSHP24-3 and pHNSHP24-4 which were bound to two copies of IS26. (Figure 2). In addition, the fusion plasmid pHNSHP24 was both identified in the transconjugant and the parental strain SHP24. Thus, the hybrid plasmid pHNSHP24 carrying mcr-1.1 may be formed before conjugation and may be the fusion product of pHNSHP24-3 and pHNSHP24-4.
Based on the complete sequence analysis of the fusion plasmid and two daughter plasmids, we proposed a formation model for the fusion plasmid (Figure 2B). In this model, IS26 adjacent to the autotransporter outer membrane gene (named aomP) in pHNSHP24-4 (F53:A-:B-) recognized and attached the target site duplication (TSD) (ATCGCTTA) in the non-conjugative plasmid pHNSHP24-3 (pO111), thus forming a replicative cointegrate intermediate. Subsequently, pHNSHP24-3 was incorporated into pHNSHP24-4, producing a cointegrated pHNSHP24, accompanied by an 8-bp direct repeat (ATCGCTTA), and an additional IS26 copy appeared upstream of the phage tail fiber gene (named ptfP).
The Conjugation Frequencies of the Fusion Plasmid pHNSHP24
The conjugation frequency of the fusion plasmid pHNSHP24 from parental strain E. coli SHP24 to recipient E. coli C600 was 2.95 ± 0.36 × 10−7, while the conjugation frequency of pHNSHP24 from the transformant E. coli DH5α/pHNSHP24 to recipient E. coli C600 was 2.66 ± 1.44×10−4.
Discussion
Multiple previous studies have reported the discovery of phages carrying multiple antibiotic resistance genes (ARGs).22,23 However, the role of phages in the transmission of antibiotic resistance genes remains a controversial problem.24 Nevertheless, some studies have shown that phages are capable of transferring resistance genes through phage-mediated transduction and promoting horizontal gene transfer of plasmids through transformation.25,26
In the present study, mcr-1.1 was found in the phage-like plasmid pHNSHP24-3 which showed highly similar to other mcr-1.1 positive phage-like plasmids from animal, human and environmental samples (Table S2), indicating that phage-like plasmids play significant vehicle role in the dissemination of mcr-1.1.8,9,27 IncI2, IncX4 and IncHI2 plasmids are predominant mcr-1.1-carrying plasmids with self-transmissible capacity, high stability and fitness advantage.5 Contrastingly, the phage-like plasmids have fitness disadvantages and were non-conjugative. However, they can be integrated with conjugative plasmids to form fusion plasmids, such as pD72C and pHNSHP24, thus acquiring conjugative transfer ability, which would accelerate the spread of resistance genes.5,7–9,28
pHNSHP24 was a cointegrate plasmid formed by an IncF53:A-:B- conjugative plasmid and a non-conjugative mcr-1.1-carrying phage-like pO111 plasmid, mediated by IS26 through a replicative transposition mechanism.29 Previous studies also reported the formation of some hybrid plasmids mediated by IS26, such as pD72C (fusion of IncN1-F33:A-:B- and phage-like pO111 plasmid), pS13D (fusion of IncN1-F33:A-:B- and IncFI:A-:B- plasmid), pSE380T (fusion of IncHI2 and virulence-carrying IncFIA plasmid), pL53T (fusion of IncX3 and IncFII plasmid), and pSL131 (fusion of IncA/C and IncX3 plasmid).7,8,30–32 It is noteworthy that the formation of cointegrate plasmids pHNSHP24, pD72C (MK419152), and pSE380T (KY401053) was because of the IS26 in the conjugative plasmids which attack the TSD of the non-conjugative plasmids.7,8,32 The fusion of plasmids enables non-conjugative plasmids to obtain self-transmissible capacity, promoting the dissemination of the ARGs carried by the non-conjugative plasmids.
The number of mcr-1.1-bearing hybrid plasmids is currently increasing (Table S1). Of the 627 mcr-1.1-bearing plasmids, 51 were hybrid plasmids, including IncHI2-IncN (n = 25), IncHI1-IncFIA (n = 7), pO111-IncFIB (n = 1), pO111-IncFII-InN (n = 1), and IncY-IncHI2-IncN (n = 1) (Table S3). These hybrid plasmids typically carry multiple antibiotic resistance genes (Table S3). Interestingly, the mcr-1.1-carrying hybrid plasmids pHNSHP24 and pD72C both were found under antimicrobial selection pressure during conjugation assays.7 The same phenomenon was also occurred in the formation of hybrid plasmids pS13D (CP047094), pC21-F1 (MT554516), and pC21-F2 (MT554517).8,14 pHNSHP24-3 (pO111) carrying mcr-1 was a non-conjugative plasmid,7,8,14 indicating the formation of the fusion plasmid pHNSHP24 (pO111-FII) might has occurred before conjugation, and we confirmed the existence of the fusion plasmids in the original strain SHP24. But the S1-PFGE and WGS data of the original strain SHP24 failed to detect the fusion plasmid, indicating the abundance of the fusion plasmid in SHP24 is low. During conjugation experiments, fusion plasmids transferred to the recipient and transconjugants carrying fusion plasmids may be enriched under the antimicrobial selective pressure. In contrast, some previous studies suggested that the fusion plasmid may be formed during conjugation.31,33 Thus, the fusion plasmid formed before or during conjugation still remain controversial. Nevertheless, it seemed that the antimicrobial pressure promoted the selection and dissemination of fusion plasmids. Therefore, we must charily use antibiotic drugs to avoid the emergence and transmission of fusion plasmids.
Conclusion
This study firstly reported the fusion of an IncF53:A-:B- conjugative plasmid with a non-conjugative mcr-1.1-bearing phage-like pO111 plasmid mediated by IS26. The formation of conjugative fusion plasmids may stimulate the spread of mcr-1.1 and other resistance genes, and more attention should be paid to fusion plasmids derived from phage-like plasmids and conjugative plasmids.
Acknowledgments
This work was supposed in part by the National Natural Science Foundation of China (No. 31830099 and 32141002), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2019BT02N054), and the Innovation Team Project of Guangdong University (No. 2019KCXTD001).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
2. Wang R, van Dorp L, Shaw LP, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9(1):1179. doi:10.1038/s41467-018-03205-z
3. Matamoros S, van Hattem JM, Arcilla MS, et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. 2017;7(1):15364. doi:10.1038/s41598-017-15539-7
4. Sun J, Zhang H, Liu YH, Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26(9):794–808. doi:10.1016/j.tim.2018.02.006
5. Wu R, Yi LX, Yu LF, et al. Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front Microbiol. 2018;9:331. doi:10.3389/fmicb.2018.00331
6. Ogura Y, Ooka T, Iguchi A, et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A. 2009;106(42):17939–17944. doi:10.1073/pnas.0903585106
7. He D, Zhu Y, Li R, et al. Emergence of a hybrid plasmid derived from IncN1-F33: a-:B-and mcr-1-bearing plasmids mediated by IS26. J Antimicrob Chemother. 2019;74(11):3184–3189. doi:10.1093/jac/dkz327
8. Liu YY, He DD, Zhang MK, et al. The formation of two hybrid plasmids mediated by IS26 and Tn6952 in salmonella enterica serotype enteritidis. Front Microbiol. 2021;12:676574. doi:10.3389/fmicb.2021.676574
9. Zhou W, Liu L, Feng Y, Zong Z. A P7 phage-like plasmid carrying mcr-1 in an ST15 Klebsiella pneumoniae clinical isolate. Front Microbiol. 2018;9:11. doi:10.3389/fmicb.2018.00011
10. Che Y, Xu X, Yang Y, et al. High-resolution genomic surveillance elucidates a multilayered hierarchical transfer of resistance between WWTP- and human/animal-associated bacteria. Microbiome. 2022;10(1):16. doi:10.1186/s40168-021-01192-w
11. Munby M, Fujiki J, Aoki K, et al. Whole-genome sequence of fluoroquinolone-resistant Escherichia coli HUE1, isolated in Hokkaido, Japan. Microbiol Resour Announc. 2020;9(46):e01135–e01120. doi:10.1128/MRA.01135-20
12. Shin J, Ko KS, Plasmid A. Bearing the bla(CTX-M-15) gene and Phage P1-like sequences from a sequence type 11 Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2015;59(10):6608–6610. doi:10.1128/AAC.00265-15
13. Lv LC, Lu YY, Gao X, et al. Characterization of NDM-5-producing Enterobacteriaceae isolates from retail grass carp (Ctenopharyngodon idella) and evidence of bla NDM-5-bearing IncHI2 plasmid transfer between ducks and fish. Zool Res. 2022;43(2):255–264. doi:10.24272/j.issn.2095-8137.2021.426
14. Pan Y, Zhang T, Yu L, et al. IS1294 reorganizes plasmids in a multidrug-resistant Escherichia coli strain. Microbiol Spectr. 2021;9(2):e0050321. doi:10.1128/Spectrum.00503-21
15. Shan X, Li XS, Wang N, et al. Studies on the role of IS1216E in the formation and dissemination of poxtA-carrying plasmids in an Enterococcus faecium clade A1 isolate. J Antimicrob Chemother. 2020;75(11):3126–3130. doi:10.1093/jac/dkaa325
16. Xie M, Chen K, Ye L, et al. Conjugation of virulence plasmid in clinical Klebsiella pneumoniae strains through formation of a Fusion Plasmid. Adv Biosyst. 2020;4(4):e1900239. doi:10.1002/adbi.201900239
17. Sun J, Yang RS, Zhang Q, et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol. 2016;1:16176. doi:10.1038/nmicrobiol.2016.176
18. Lucas M, Gonzalez-Perez B, Cabezas M, Moncalian G, Rivas G, de la Cruz F. Relaxase DNA binding and cleavage are two distinguishable steps in conjugative DNA processing that involve different sequence elements of the nic site. J Biol Chem. 2010;285(12):8918–8926. doi:10.1074/jbc.M109.057539
19. Wick RR, Judd LM, Gorrie CL, Holt KE, Phillippy AM. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.1005595
20. Arndt D, Grant JR, Marcu A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–W21. doi:10.1093/nar/gkw387
21. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi:10.1093/bioinformatics/btr039
22. Kasman LM. Barriers to coliphage infection of commensal intestinal flora of laboratory mice. Virol J. 2005;2:34. doi:10.1186/1743-422X-2-34
23. Rolain JM, Fancello L, Desnues C, Raoult D. Bacteriophages as vehicles of the resistome in cystic fibrosis. J Antimicrob Chemother. 2011;66(11):2444–2447. doi:10.1093/jac/dkr318
24. Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, Petit MA. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 2017;11(1):237–247. doi:10.1038/ismej.2016.90
25. Keen EC, Bliskovsky VV, Malagon F, et al. Novel “Superspreader” bacteriophages promote horizontal gene transfer by transformation. mBio. 2017;8(1):e02115–e02116. doi:10.1128/mBio.02115-16
26. Ross J, Topp E, Schaffner DW. Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction. Appl Environ Microbiol. 2015;81(22):7905–7913. doi:10.1128/AEM.02363-15
27. Li XP, Sun RY, Song JQ, et al. Within-host heterogeneity and flexibility of mcr-1 transmission in chicken gut. Int J Antimicrob Agents. 2020;55(1):105806. doi:10.1016/j.ijantimicag.2019.09.010
28. Bai L, Wang J, Hurley D, et al. A novel disrupted mcr-1 gene and a lysogenized phage P1-like sequence detected from a large conjugative plasmid, cultured from a human atypical enteropathogenic Escherichia coli (aEPEC) recovered in China. J Antimicrob Chemother. 2017;72(5):1531–1533. doi:10.1093/jac/dkw564
29. He S, Hickman AB, Varani AM, et al. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio. 2015;6(3):e00762. doi:10.1128/mBio.00762-15
30. Li R, Xie M, Liu L, et al. Characterisation of a cointegrate plasmid harbouring blaNDM-1 in a clinical Salmonella Lomita strain. Int J Antimicrob Agents. 2020;55(1):105817. doi:10.1016/j.ijantimicag.2019.09.021
31. Liu Z, Xiao X, Liu Y, Li R, Wang Z. Recombination of NDM-5-producing plasmids mediated by IS26 among Escherichia coli. Int J Antimicrob Agents. 2020;55(1):105815. doi:10.1016/j.ijantimicag.2019.09.019
32. Wong MH, Chan EW, Chen S. IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. J Antimicrob Chemother. 2017;72(10):2750–2754. doi:10.1093/jac/dkx238
33. Xie M, Li R, Liu Z, Chan EWC, Chen S. Recombination of plasmids in a carbapenem-resistant NDM-5-producing clinical Escherichia coli isolate. J Antimicrob Chemother. 2018;73(5):1230–1234. doi:10.1093/jac/dkx540
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.